FIGURE 1.
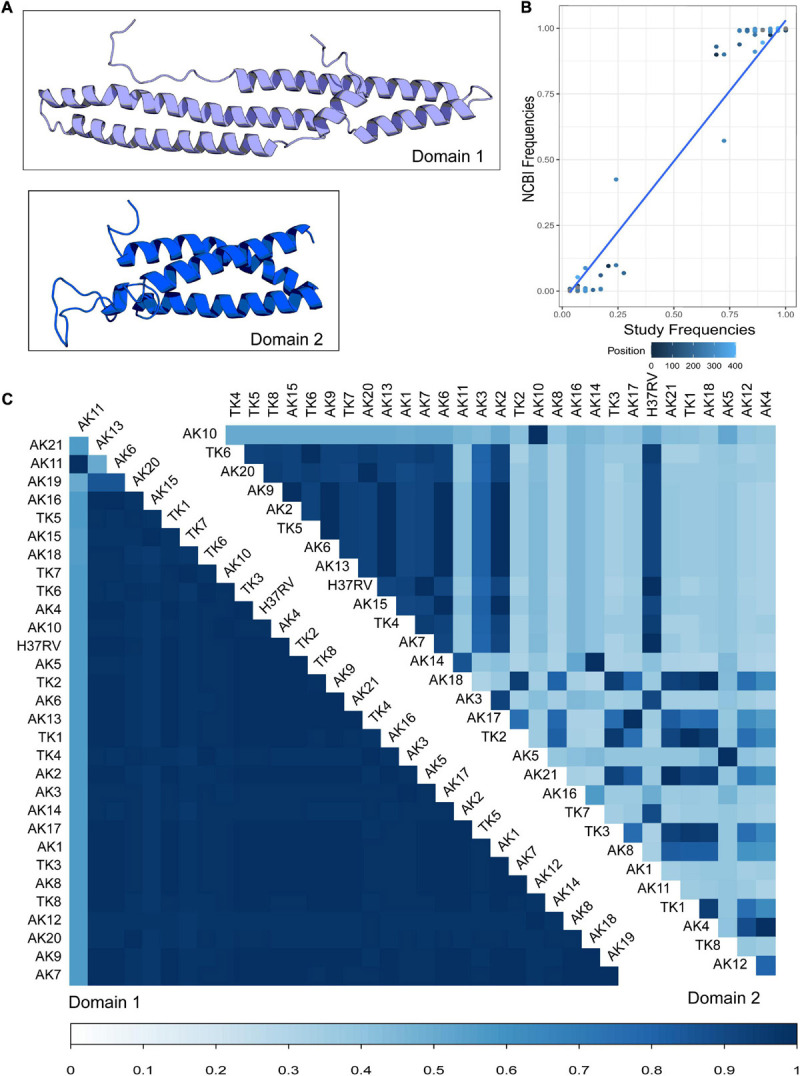
Modeling and comparison of Mycobacterium tuberculosis (MTB) protein PPE18 structures of clinical strains. (A) Predicted structures of two PPE18 protein domains for MTB laboratory reference strain H37Rv using RaptorX template-based tertiary structure prediction. (B) Comparison of the frequencies of the most common residue at each position in the PPE18 protein sequence between the 30 sequences modeled in this study and 1,438 complete PPE18 sequences obtained from the NCBI protein database. Each dot represents a given common residue between the two groups of sequences. Color indicates position in the sequence. Blue line shows a fitted GLM with P-value <2E-16. (C) Pairwise comparison of TM-align scores for variant strains investigated in this study. Variants AK1–AK21 are sequences found in Arkansas MTB strains. Variants TK1–TK8 represent sequences found in the Turkey MTB strains. A TM-align score of 0.3 and lower indicates random structural similarity. A TM-align score of 0.5 and higher indicates the same fold of similarity between the protein structures in comparison. Bottom bar indicates TM-align score.
