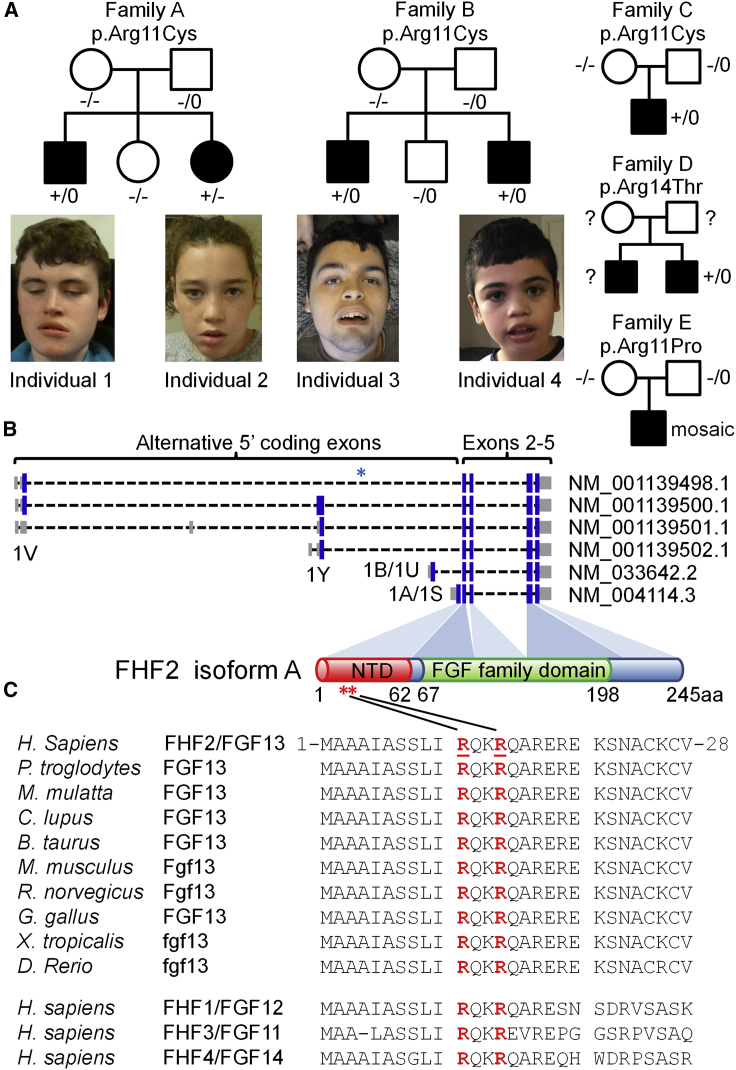
Figure 1.
Family pedigrees and locations of the FHF2A variants
(A) Pedigrees of families A–E with photographs of the affected individuals from families A and B. Genotypes are heterozygous mutant (+/−), homozygous wild type (−/−), hemizygous mutant (+/0), hemizygous wild type (−/0), or unknown (?).
(B) Exonic models of the FHF2 transcripts showing the alternative 5′ coding exons. The FHF2A protein isoform is derived from the GenBank: NM_004114.5 transcript which is initiated from an ATG start site in exon 1A (also known as exon 1S). The V, Y, U, and S nomenclature originates from Munoz-Sanjuan et al.27 The 1A/1S exon encodes the N-terminal domain (NTD) of FHF2A (amino acid residues 1 to 62). The shared C-terminal exons encode the core fibroblast growth factor (FGF) domain (amino acid residues 67 to 198). The blue asterisk indicates the location of the breakpoint of the translocation described by Puranam et al.28 The red asterisks indicate the location of the DEE-associated missense variants.
(C) The FHF2 (FGF13) substitutions are located at highly conserved residues in the N-terminal domain of the A isoform. Homology alignments for human FHF2 (GenBank: NP_004105.1, amino acid residues 1–28) and a range of orthologs and paralogs. Orthologs include chimpanzee (GenBank: XP_001138460.1), rhesus macaque (GenBank: NP_001252772.1), dog (GenBank: XP_549294.2), cow (GenBank: NP_001092362.1), mouse (GenBank: NP_034330.2), rat (GenBank: XP_006257654.1), chicken (GenBank: XP_015133693.1), western clawed frog (GenBank: XP_012824080.1), and zebrafish (GenBank: XP_005173268.1). Paralogs include FHF1/FGF12 (GenBank: NP_066360.1), FHF3/FGF11 (GenBank: NP_004103.1), and FHF4/FGF14 (GenBank: NP_004106.1). The positions of the Arg11 and Arg14 residues are underlined and in red.