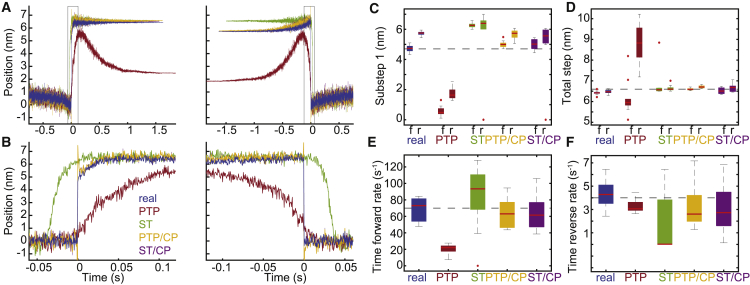
Figure 5.
Ensemble averages of simulated binding interactions. In total, 10 sets of data were simulated, each containing 100 binding interactions (sets 1–10). Interactions were detected using either the peak-to-peak (PTP) or the single-threshold (ST) method, and interactions were aligned using either the transitions estimated by the covariance threshold method or the change points identified by the change-point algorithm (CP). (A) (left) For each analysis method, all detected binding interactions were aligned at the estimated initiation times and averaged together to generate time-forward ensemble averages. (right) For each analysis method, all detected binding interactions were aligned at the estimated termination times and averaged together to generate time-reversed ensemble averages. Also shown are the time-forward and time-reversed ensemble averages generated from the known locations of the actual simulated binding interactions (blue, real). (B) A zoomed-in view of the boxed segments of the ensemble averages in (A) highlights the misalignment in the averages when the change-point algorithm is omitted. (C–F) For each of the 10 simulated sets of data containing 100 binding interactions, ensemble averages were generated and fit with single exponential functions. The substep sizes and rates of the simulated myosin working stroke were estimated from the exponential fits. Box plots show the estimated parameters for each analysis method. Outliers are indicated by red dots. The substep sizes were estimated from both the time-forward (f) and the time-reversed (r) ensemble averages. Horizontal dashed lines show the values of the simulated parameters. Statistical analysis for each parameter can be found in Table S3. To see this figure in color, go online.