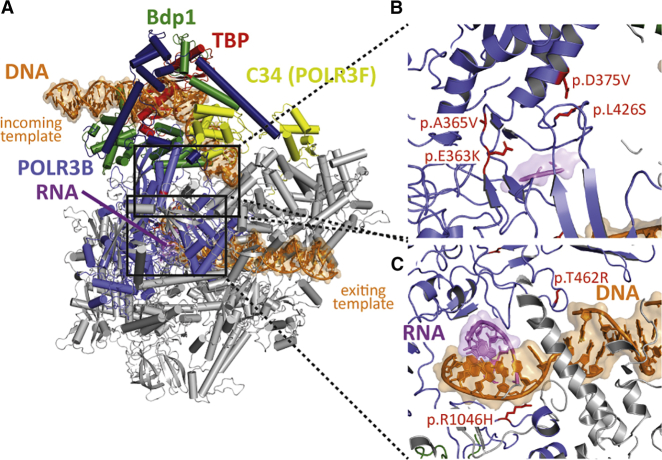
Figure 1.
Structural protein modeling
(A) In order to localize the POLR3B variants found in the patients on the yeast POLR3B structure, we used the model of POLR3 in transcription initiation state.13
(B) Of the six variants identified, four of them cluster in a region of POLR3B where transcribed DNA is melted.
(C) No direct contacts with other POLR3 subunits were identified, but the mutated region is at the transcription bubble and may have an impact on transcription itself. Similarly, the two remaining variants are located in the exiting DNA tunnel and have direct contact with the duplex DNA. These variants may affect transcription activity. Of note, none of the identified variants are at the interface with transcription factors or transcription repressors (e.g., Maf1). POLR3B-altered residues found in the six patients and their equivalent yeast positions are as follows: Asp375 = Asp390, Leu426 = Leu444, Arg1046 = Arg1061, Ala365 = Ala380, Glu363 = Glu378, and Thr462 = Ser480.