Figure 1.
Genotype and Erythroid Phenotype of Individuals with VPS4A Mutations
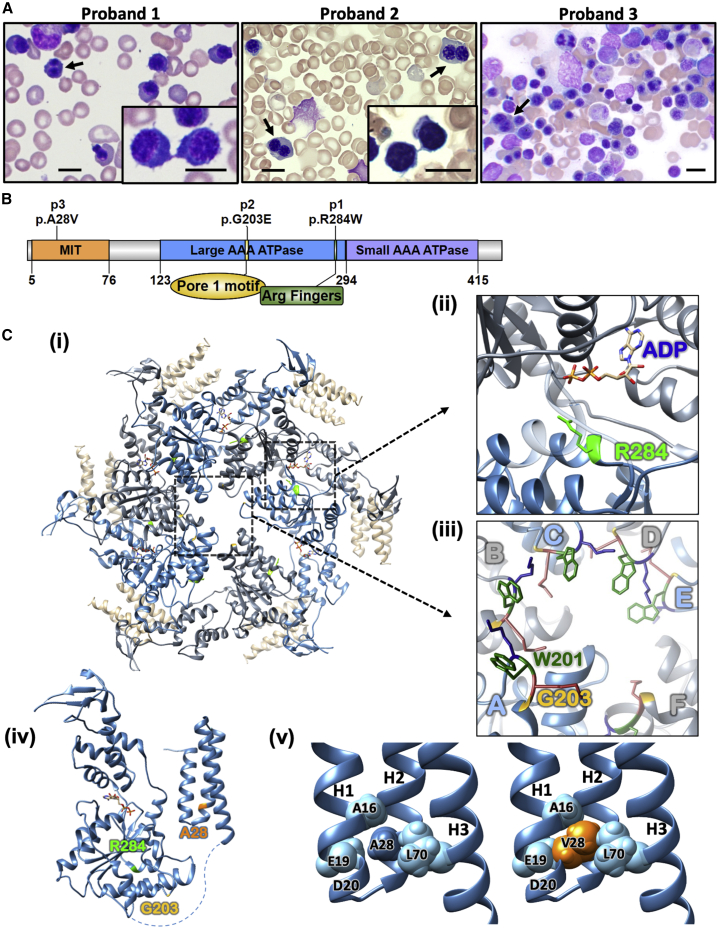
(A) Bone marrow aspirate smears from each proband showing erythroid hyperplasia. Erythroblasts show megaloblastoid changes and include cells with binucleation (arrows) and cytoplasmic bridges joining erythroblasts post-division, noted especially in probands 1 and 2 (insets). Scale bar 10 μm.
(B) Diagram of VPS4A protein structure. The N-terminal MIT domain is followed by a flexible linker and an ATPase cassette consisting of a large and small ATPase domain and a β-domain. The location of the mutations found in the probands is marked.
(C) (i) Ribbon diagram of Vps4 proteins assembled in a hexamer (PDB: 6AP1). Residues mutated in the probands are colored green (Arg284 [R284] in p1) and yellow (Gly203 [G203] in p2). The numbering for human VPS4A is used, depicting the homologous residues in the yeast Vps4. Insets show (ii) the ADP in binding site at the interface of two adjacent subunits (blue and gray) with Arg284 (R284 in green) and (iii) the central pore of the hexamer where Gly203 (G203 in yellow) in the pore loop is crucial for the arrangement of Trp201 (W201 in dark green) forming hydrophobic binding pockets for ESCRT-III proteins passing through the central pore.12 (iv) Ribbon diagram of a Vps4 monomer based on PDB: 6AP1 for yeast Vps4 and PDB: 1YXR for human MIT domain. Residues mutated in the probands are colored green (Arg284 [R284] in p1), yellow (Gly203 [G203] in p2), and orange (Ala28 [A28] in p3). (v) The structure of human VPS4A MIT domain (PDB: 1YXR) with Ala28 (A28) and neighboring residues shown as spheres (left). Modeling of the Val28 variant (orange) found in p3 demonstrates clashes with the side chains of Leu70 and Glu19, as well as the main chain of Ala16 and Asp20 (side chain not shown for clarity) (right). All protein structure figures were generated with UCSF Chimera.13