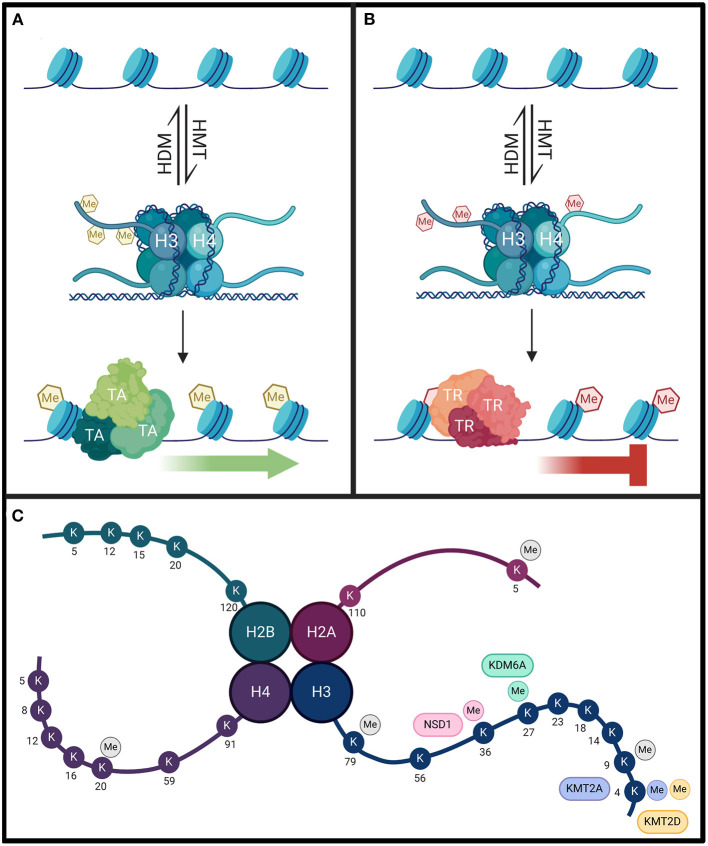
Figure 1.
Histone methylation regulates gene expression through recruitment of different transcription factors rather than by directly altering chromatin structure. (A) When histone methyltransferases (HMT) methylate lysine or arginine residues on H3 and H4 associated with active chromatin (yellow hexagons), transcriptional activators (TA) can be recruited to those sites to promote gene expression. (B) However, when HMTs target different residues on H3 and H4 that are associated with chromatin repression, these methylated regions (red hexagons) can recruit transcriptional repressors to silence the gene. The transcriptional effects of histone methylation can be reversed by histone demethylases (HDM). Proper gene expression is dependent on the homeostasis of HMT and HDM enzyme activity. (C) Lysine methylation occurs primarily on H3 and H4. Each KMT/KDM will only target select residues, and the enzymes discussed have high specificity for these targets. Mutations that impair the function of these enzymes would be expected to affect the methylation status of these residues. Each enzyme and their lysine methylation sites are color coded. Other lysine methylation sites not specifically targeted by these enzymes are indicated in gray. Arginine methylation sites are not indicated in this figure.