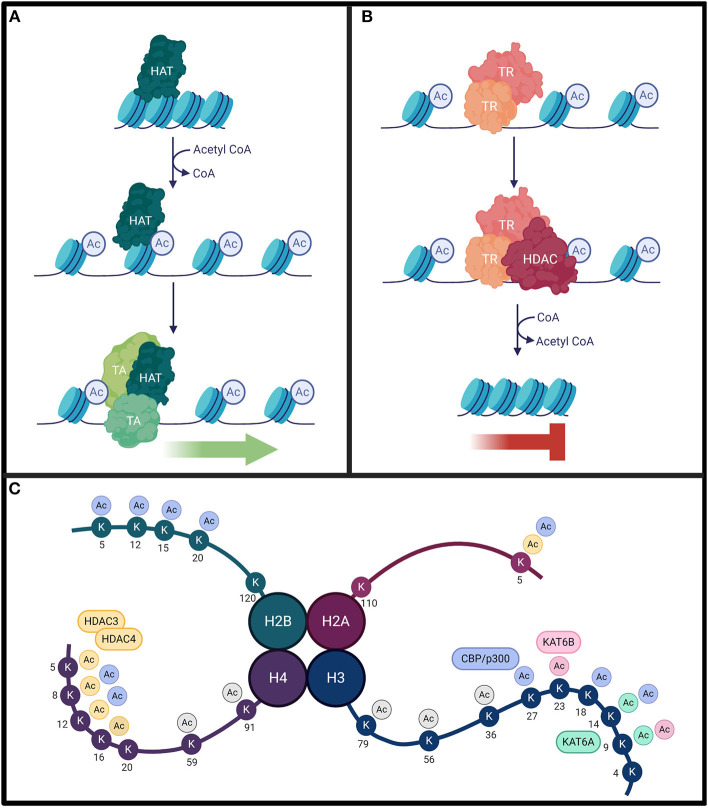
Figure 2.
(A) Histone acetyltransferases (HAT) will acetylate histone lysine residues using acetyl CoA cofactor (blue circles). This weakens the electrostatic interactions between positively-charged histones and negatively-charged DNA to loosen chromatin structure. This results in DNA exposure, allowing for the recruitment of transcriptional activators (TA). (B) Conversely, transcriptional repressor (TR) complexes can interact with histone deacetylases (HDAC) to remove these modifications. This strengthens the electrostatic interactions between the DNA and the histones, resulting in compact chromatin, inhibiting transcription. The homeostasis between histone acetylation and deacetylation is critical for proper gene expression. (C) Lysine acetylation can occur on all four histone subunits. Each HAT/HDAC has preferred target sites. While KAT6A and KAT6B acetylate residues with high specificity, CBP and HDAC4/HDAC3 complexes have a broader range of targets. The lysine residues targeted by each enzyme are color coded. There is some target redundancy between HAT/HDACs, and other histone (de)acetylating enzymes can also target common sites (not indicated on figure). Additional lysine acetylation sites not specifically targeted by these enzymes are indicated in gray.