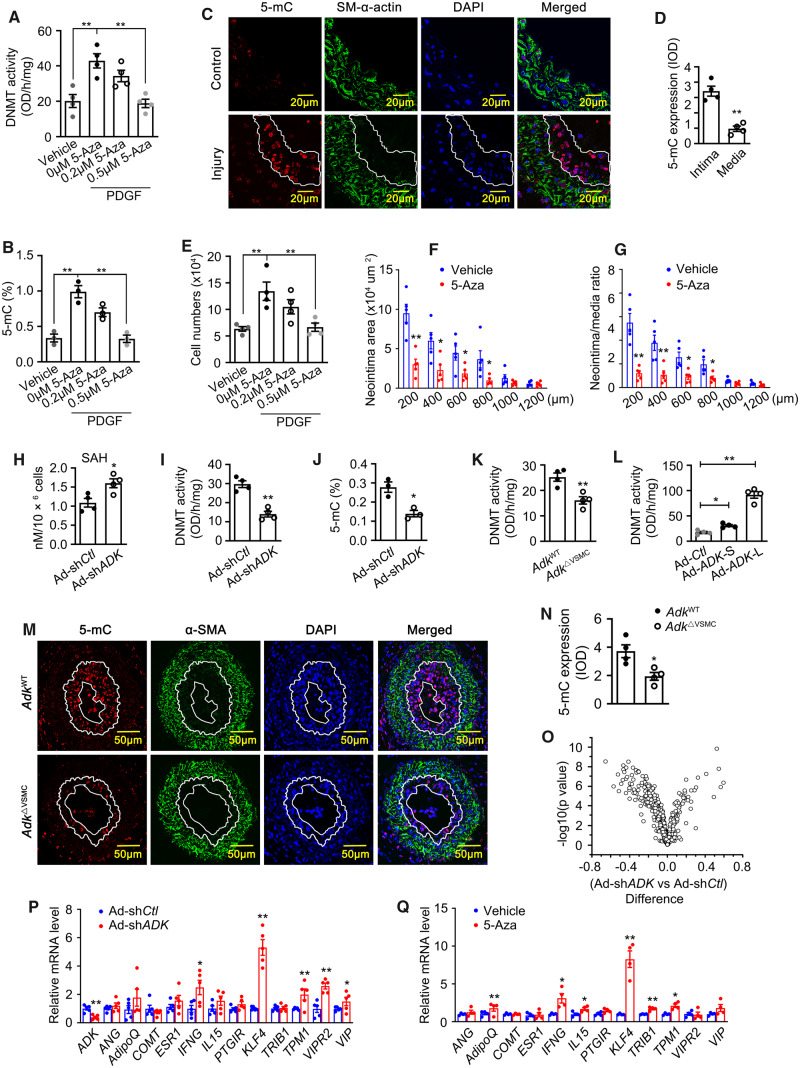
Figure 4.
Involvement of ADK-driven DNA hypermethylation in VSMC proliferation and neointima formation. Quantification of DNA methyltransferase (DNMT) activity (A) and 5-methylcytosine (5-mC) content (B) in HCSMCs pretreated with 5-Aza (2 μmol/L or 5 μmol/L) for 1 h and followed by incubation with PDGF (20 ng/mL) for 72 h (n = 4 for A and 3 for B). Representative IF staining (C) and quantification (D) of 5-mC on sections of mouse carotid arteries from mice without or with ligation injury (n = 4 mice per group). Arterial neointima is indicated within the white line. (E) WST-1 proliferation analysis of HCSMCs pretreated with 5-Aza-2′-deoxycytidine (5-Aza, 2 μmol/L or 5 μmol/L) for 1 h followed by incubation with PDGF (20 ng/mL) for 72 h (n = 4). Quantitative analysis of arterial neointima area (F) and ratios of neointima areas to medial areas of injured carotid arteries (G) from 5-Aza-treated mice (n = 5 mice per group). Quantification of SAH level (H), DNMT activity (I), and 5-mC content (J) in control and ADK KD HCSMCs growing in complete medium (n = 3 or 4 independent experiments). Quantification of DNMT activity (K) in mouse aortic SMCs isolated from AdkWT or AdkΔVSMC mice (n = 4 independent experiments). (L) Quantification of DNMT activity in HCSMCs transfected with the ADK-S and ADK-L isoforms (n = 4 independent experiments). Representative IF staining (M) and quantification of 5-mC (N) on sections of mouse carotid arteries from AdkWT or AdkΔVSMC mice with ligation injury. The area highlighted with the white line is the arterial neointima (n = 4 mice per group). (O) Mean methylation difference of CpG sites in the promoters of 30 anti-proliferative genes of VSMCs comparing control group with those of ADK KD HCSMCs. n = 3 independent cultures, x axis: mean methylation difference of CpG sites between control and ADK KD HCSMCs; y axis: −log10 of the P-values. (P) Quantitative RT–PCR analysis of mRNA levels for anti-proliferative genes with DNA hypomethylation in the promoters due to ADK KD in control and ADK KD HCSMCs (n = 6 independent experiments). (Q) Quantitative RT–PCR analysis of mRNA levels for genes in (H) in HCSMCs treated with 5 μmol/L 5-Aza or vehicle for 72 h (n = 4 or 5 independent experiments). For all bar graphs, data are expressed as the means ± SEM, *P < 0.05 and **P < 0.01 (one-way ANOVA with Tukey’s post hoc test for A, B, E, and L; unpaired, two-tailed Student’s t-test for D, F–K, and N–Q).