Figure 1.
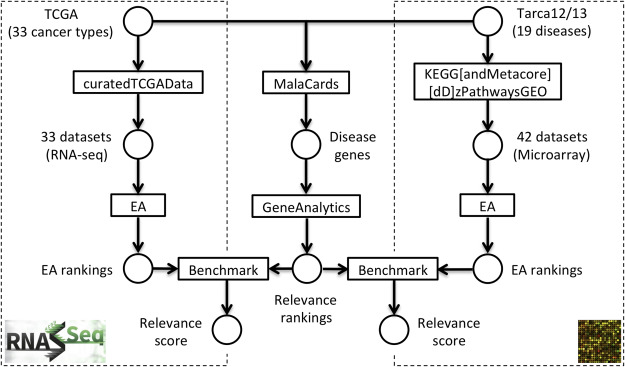
Benchmark setup. The benchmark framework incorporates a pre-defined RNA-seq panel (left), gene set relevance rankings (center) and a microarray panel (right). The RNA-seq panel investigates 33 cancer types across 33 datasets from TCGA [32], which are accessed through the curatedTCGAData package. The microarray panel investigates 19 human diseases across 42 datasets collected by Tarca et al. [25, 26], which are available in the KEGGdzPathwaysGEO and KEGGandMetacoreDzPathwaysGEO packages. Gene set relevance rankings for both data panels are constructed by (i) querying the MalaCards database [33] for each disease investigated and (ii) subjecting resulting disease genes to GeneAnalytics [34], which yields relevance rankings for GO-BP terms and KEGG pathways. EA methods selected for benchmarking are carried out across datasets of the data panels, yielding a gene set ranking (EA ranking) for each method on each dataset. The resulting EA rankings for each dataset are then benchmarked against the precompiled relevance rankings for the corresponding disease investigated.
