Figure 1.
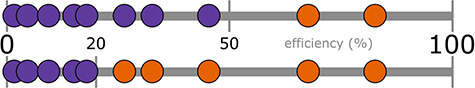
This figure represents two potential decision thresholds (20% and 50%) for 10 hypothetical sgRNA samples (colored dots). Each sample has a DNA cleavage efficiency in the range of 0% and 100%. For a binary classifier, samples above the decision threshold are considered ‘high-efficiency’ (orange) and samples below the decision threshold are considered ‘low-efficiency’ (purple). The decision threshold can be arbitrarily set to any value between 0% and 100%, and an appropriate decision threshold can help keep data balanced. A threshold of 50% in the upper example results in two ‘highs’ and eight ‘lows’. This can result in a poor-performing model as a resulting model could indiscriminately classify all 10 targets as low-efficiency (purple) yet have a relatively good accuracy of 80% (8 out of 10 are correct). However, a threshold of 20% results in five highs and five lows. Now if a model indiscriminately classifies all 10 targets as low-efficiency it will have a more appropriate accuracy of 50% hence being forced to learn the discriminating features between the two classes.
