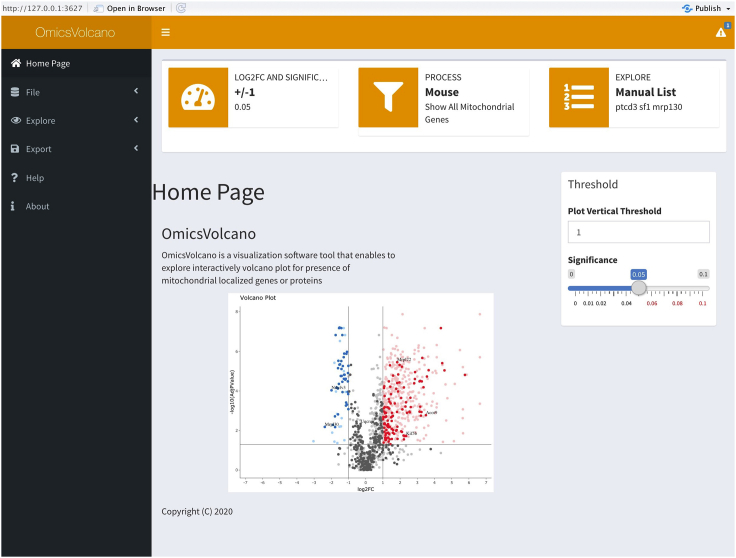
Figure 1.
The OmicsVolcano home page
The software requires four steps to generate the interactive omics-data visualization plots. The “file” option allows the upload of the input data as an ASCII file which contains IDs, gene names, gene descriptions, log2 fold changes, and adjusted p values. The “explore” option provides several main core functionalities of the software: plot, custom gene or protein selection, mitochondrial processes, multiple process visualization, and cellular compartment visualization. Customization of the statistical significance and threshold of the y-axis, are performed by adjustments of the slider widgets. Additional options, e.g., “upload a gene file”, “insert a list of genes”, “select organism”, and “show mitochondrial process” widgets allow the customization of the data exploration processes. The “export” option enables the export of data as tables or graphics in various pre-defined formats. A manual is available through the “help” option, and the software package version including additional information about the software can be found in the “about” option.