Fig. 1.
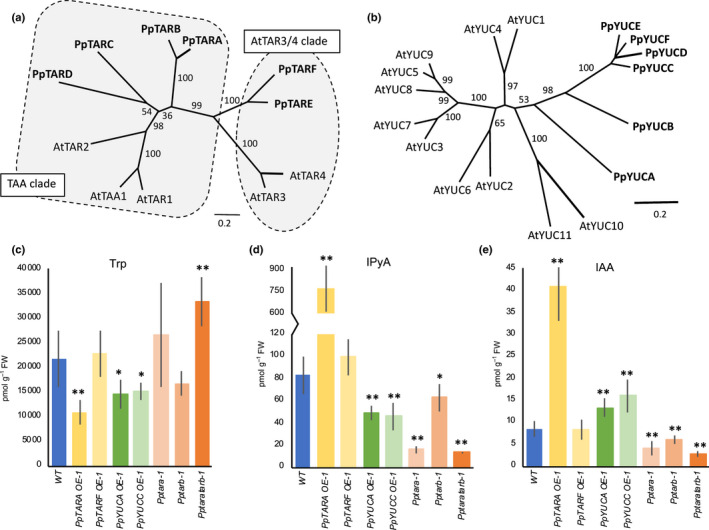
TAR and YUC multigene families promote auxin biosynthesis in Physcomitrium (Physcomitrella) patens. (a, b) Unrooted phylogenetic trees produced by the maximum likelihood method to infer evolutionary relationship of (a) TAR and (b) YUC proteins encoded by Arabidopsis (At) and P. patens (Pp) genes. Branch lengths indicate genetic distance and numbers on branches are bootstrap values (only values above 50% are shown). The TAR proteins fall in two distinct clusters referred to as the TAA and the AtTAR3/4 clades, respectively. (c–e) Level of (c) tryptophan (Trp), (d) indole‐3‐pyruvic acid (IPyA) and (e) IAA in protonemal tissue of P. patens wild type (WT) and stated over‐expressor (OE) and knock‐out mutant lines. Error bars represent the SD of the mean of at least five biological replicates. Asterisks indicate statistically significant differences compared to WT: *, P < 0.05; **, P < 0.01 (Student’s t‐test). See also Supporting Information Table S3.
