Fig. 2.
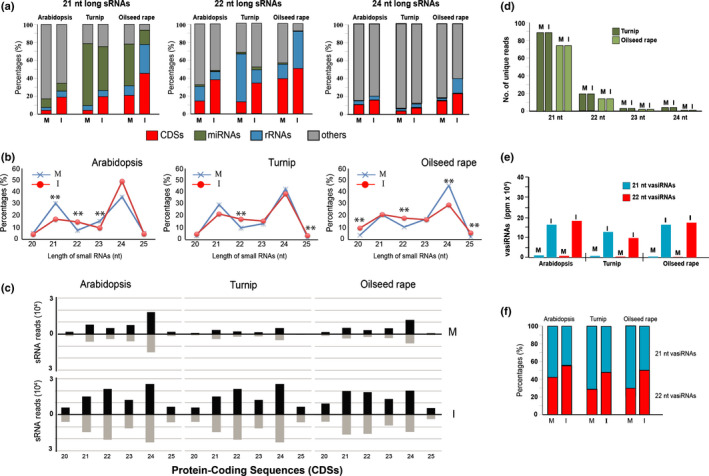
Origin of small RNAs (sRNAs) in Arabidopsis thaliana, turnip, and oilseed rape infected by cauliflower mosaic virus (CaMV) and 21 nt and 22 nt virus‐activated small interfering RNA (vasiRNA) in infected plants. (a) Incidence of 21, 22, and 24 nt total sRNAs according to their different origin. (b) Profile of unique sRNAs according to their lengths, with statistical evaluation of changes (**, adjusted P < 0.01, χ 2 test). (c) Length distribution and abundance (reads per million × 104) of the sRNAs unique reads derived from protein‐coding sequences (CDSs) in mock‐treated (M) or CaMV‐infected (I) plants. (d) Size distribution profile of sRNAs from microRNA (miRNA) precursor transcripts. (e) The 21 nt and 22 nt unique vasiRNAs from 15 orthologous transcripts. The values are given in parts per million (ppm) referring to all unique 20–25 nt sRNAs. (f) Ratio of 21 nt and 22 nt vasiRNAs (21 nt + 22 nt = 100). M, mock‐treated; I, CaMV infected; CDSs, protein‐coding DNA sequences; rRNAs, ribosomal RNAs; length of sRNAs is in nucleotides (nt).
