Fig. 4.
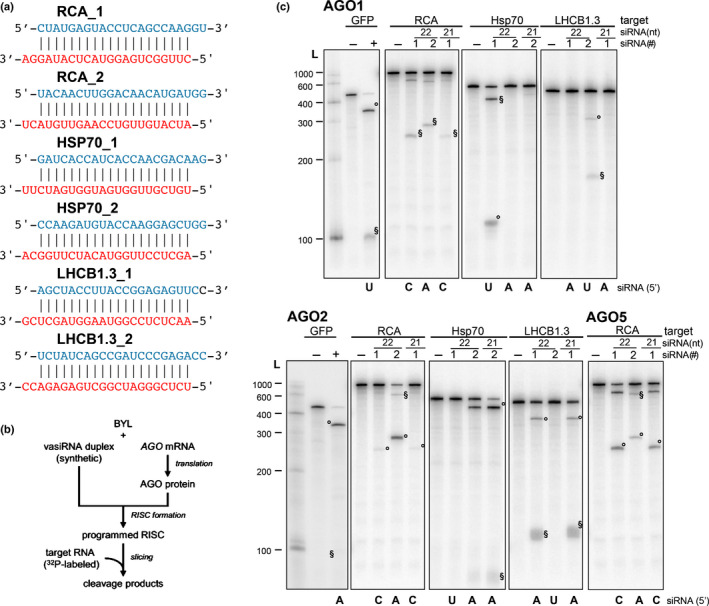
Slicer activity of 22 nt virus‐activated small interfering RNAs (vasiRNAs). (a) List of 22 nt vasiRNA duplexes used in the slicer activity assays. The guide and passenger strands are depicted in red and blue, respectively. The 21 nt long versions are shortened by one nucleotide at each strand (i.e. 3′ at the guide strand and 5′ at the passenger strand). (b) Schematic representation of the ‘slicer assay’. Argonaute (AGO)1, AGO2, or AGO5 was generated by in vitro translation in the Bright Yellow lysate (BYL) in the presence of synthetic vasiRNA duplexes generating programmed RNA‐induced silencing complexes (RISCs). Subsequently, phosphorus‐32 (32P)‐labeled, in vitro transcribed messenger RNA (mRNA) fragments containing the respective vasiRNA target sites were added and analyzed for RISC‐mediated cleavage by denaturing polyacrylamide gel electrophoresis and autoradiography. (c) Arabidopsis thaliana AGO1, AGO2, or AGO5 mRNA was translated in BYL in the presence of the individual indicated vasiRNAs to form AGO/RISC. The 5′ and 3′ cleavage products are indicated as ‘°’ and ‘§’, respectively. L, radiolabeled 100 nt RNA ladder; vasiRNAs duplexes listed in (a) are indicated with their identifying number, #1 or #2. In the negative controls (−), no small interfering RNA (siRNA) was included in the cleavage assay. Cleavage of a fragment of the mRNA of green fluorescent protein (GFP) and a GFP‐specific siRNA was used as a positive control (+).
