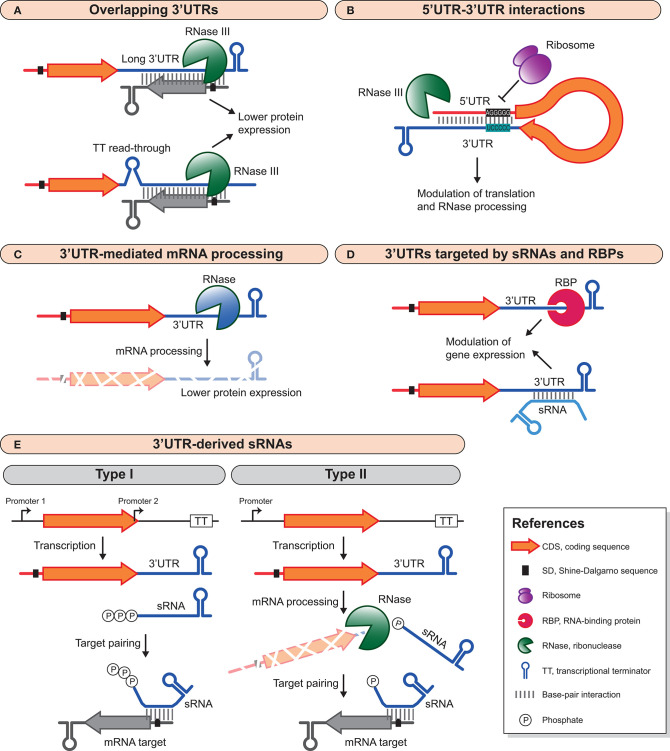
Figure 1.
Different 3′UTR-mediated regulatory mechanisms in bacteria. (A) Long 3′UTRs and transcriptional read-through can produce overlapping 3′UTRs that modulate the expression of convergent genes. These overlapping double-stranded RNA regions are processed by RNase III, which ultimately decreases protein expression. (B) 3′UTRs can interact with the 5′UTR of the same mRNA to modulate mRNA stability and translation. This interaction can inhibit translation and recruit RNases for mRNA degradation, or it can promote mRNA stability by impairing RNase processing. (C) RNases can specifically target 3′UTRs to process mRNAs and modify their half-life and protein expression yield. (D) sRNAs and RBPs can target 3′UTRs to modulate mRNA expression. This interaction can be both positive, by blocking RNase processing and enhancing mRNA stability, or negative by promoting inhibition of translation and RNase processing. (E) 3′UTRs are also reservoirs of trans-acting sRNAs than can be generated by an internal promoter within or immediately downstream of the CDS (type I) or by mRNA processing (type II). For examples, see text and Table 1.