Figure 5.
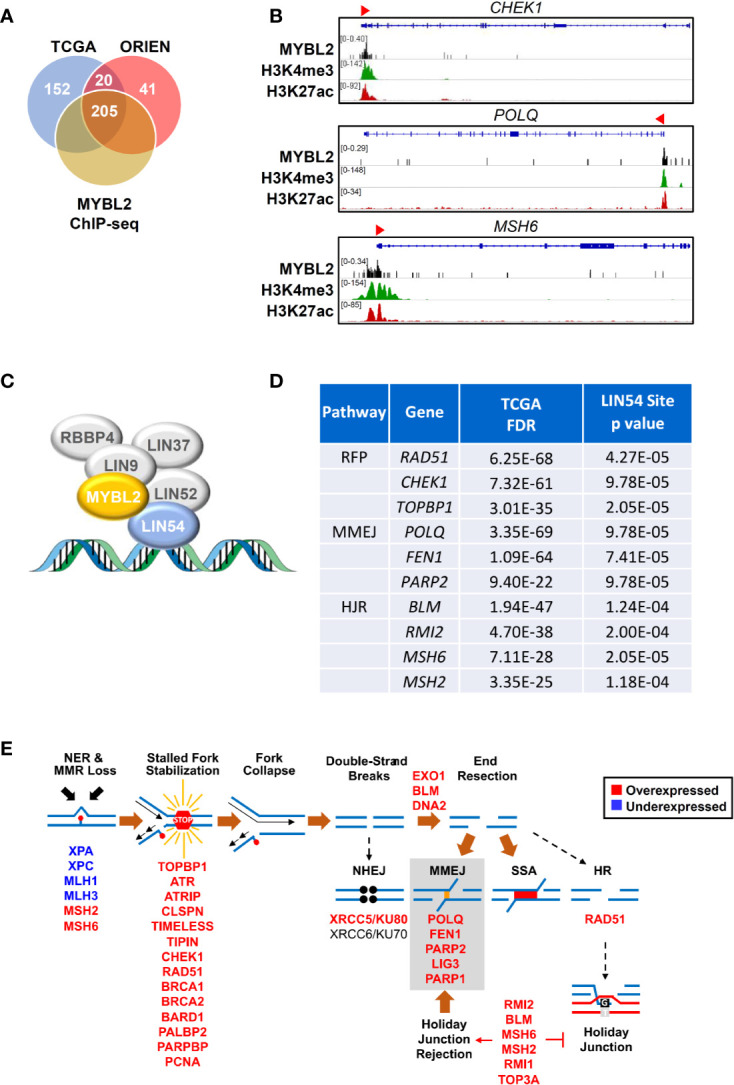
MYBL2 drives expression of genes that antagonize homologous recombination and promote error prone DNA repair. (A) Analysis of chromatin immunoprecipitation sequencing (ChIP-seq) data revealed MYBL2 localization at the promoters of 205 DNA damage response genes significantly altered in both TCGA and ORIEN MYBL2 High tumors. (B) Representative Integrated Genome Viewer (IGV) snapshots of ChIP-seq enrichment peaks showing MYBL2 and active transcription histone mark co-occupancy at promoters of key DNA damage response genes. (C) MYBL2 bound to the LIN54:MUVB complex drives transcription. (D) Key replication fork protection (RFP), microhomology-mediated end joining (MMEJ), and HJ-rejection genes contained high affinity LIN54 sites in their promoters. (E) Omics data support a model wherein MYBL2 drives expression of genes that antagonize homologous recombination and promote error-prone repair. NHEJ: non-homologous end joining, MMEJ: microhomology-mediated end joining, SSA: single-strand annealing, HR: homologous recombination. Solid brown arrows indicate dominant repair pathway fate. Solid arrow (→) and inhibition (┴) symbols indicate activation or antagonization, respectively. Hashed black arrows indicate inefficient pathways.
