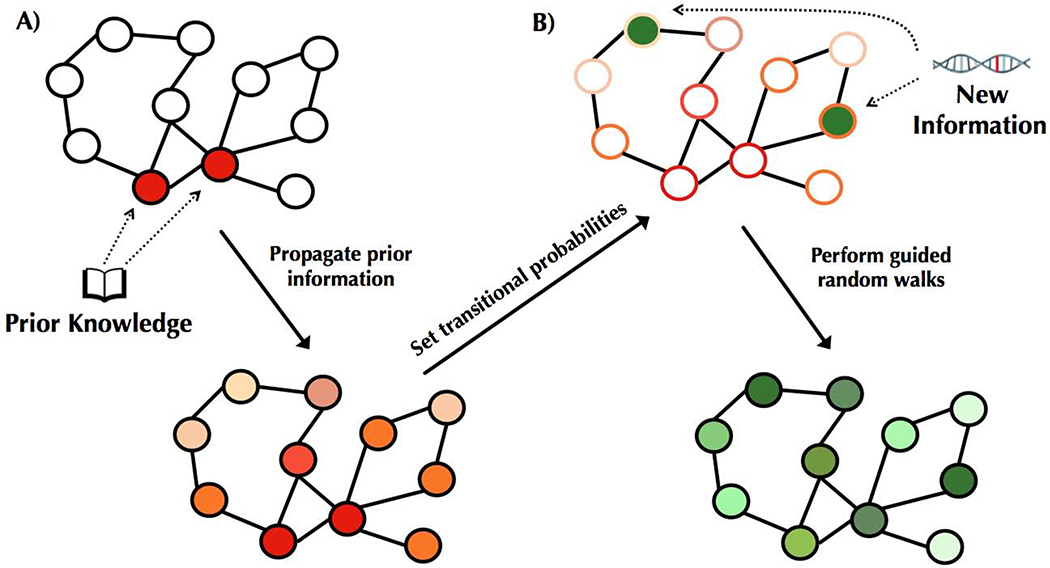
Figure 2. Overview of our approach.
(A) Known disease-relevant genes (prior knowledge) are mapped onto an interaction network (shown in red, top). Signal from this prior knowledge is propagated through the network via a diffusion approach (Qi et al., 2008), resulting in each gene in the network being associated with a score such that higher scores (visualized in darker shades of red, bottom) correspond to genes closer to the set of known disease genes. These scores are used to set transition probabilities between genes such that a neighboring gene that is closer to the set of prior knowledge genes is more likely to be chosen. (B) Genes putatively associated with the disease—corresponding to the new information—are mapped onto the network (shown in green, top). To integrate both sources of information, RWRs are initiated from the set of putatively associated genes, and at each step, the walk either restarts or moves to a neighboring gene according to the transition probabilities (i.e., walks tend to move towards genes outlined in darker shades of red). These prior-knowledge “guided” RWRs have a stationary distribution corresponding to how frequently each gene is visited, and this distribution is used to order the genes. Higher scores correspond to more frequently visited genes (depicted in darker greens, bottom).