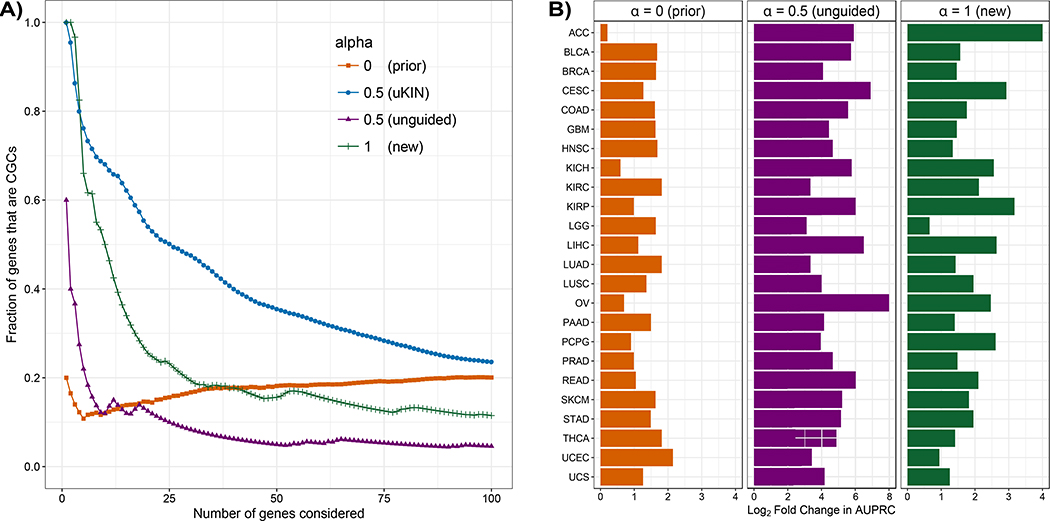
Figure 3. uKIN successfully integrates new information and prior knowledge.
(A) We illustrate the effectiveness of our approach uKIN on the GBM data set and the HPRD protein-protein interaction network using 20 randomly drawn CGCs to represent the prior knowledge. We combine prior and new knowledge using a restart probability of α = 0.5 (blue line). As we consider an increasing number of high scoring genes, we plot the fraction of these that are part of the hidden set of CGCs. As baseline comparisons, we also consider versions of our approach where we use only the new information (α = 1) and order genes by their mutational frequency (green line); where we use new information to perform unguided random walks with α = 0.5 and order genes by their probabilities in the stationary distribution of the walk (which uses new information but not prior information, purple line); and where we use only prior information (α = 0) and order genes based on information propagated from the set of genes comprising our prior knowledge (orange line). Integrating both prior and new sources of information results in better performance. (B) The performance of uKIN when integrating information at α = 0.5 is compared to the three baseline cases where either only prior information is used (α = 0, left) or when only new information is used (α = 1, right and unguided RWRs with α = 0.5, middle). In all three panels, for each cancer type, we plot the log2 ratio of the AUPRC of uKIN with guided RWRs with α = 0.5 to the AUPRC of the other approach. Across all 24 cancer types, using both sources of information outperforms using just one source of information.