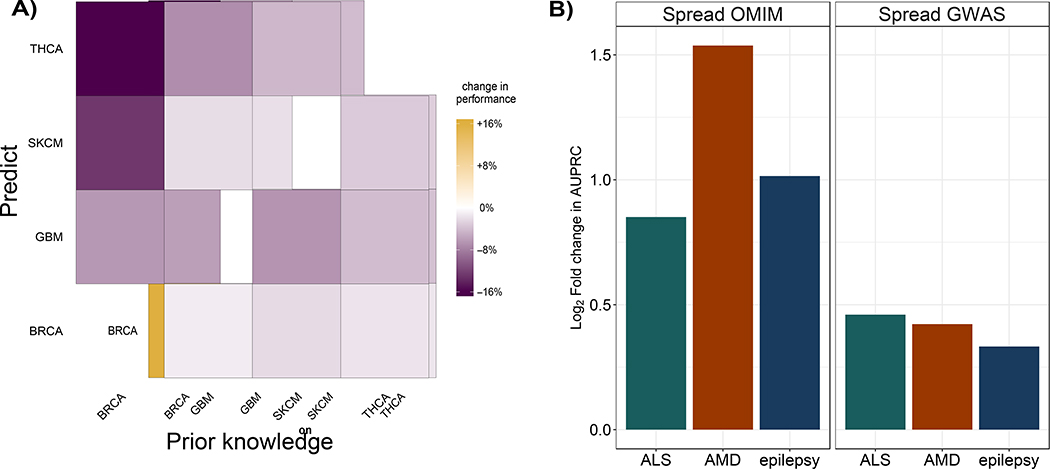
Figure 5.
(A) Use of cancer-type specific knowledge improves performance. For four cancer types, BRCA, GBM, SKCM, and THCA, we consider the performance of uKIN with α = 0.5 when using TCGA mutational data for that cancer type with prior knowledge consisting of genes known to be driver in that cancer type, as compared to performance when the prior knowledge set consists of genes that are annotated as driver only for one of the other three cancer types. For each cancer, performance is measured by the average ranking by uKIN of genes known to be driver for that cancer. For all combinations of possible prior knowledge sets (x-axis) and specific cancer gene sets that we wish to recover (y-axis), using prior knowledge from another cancer (off diagonal entries) leads to a decrease in performance as compared to the corresponding pairs (diagonal entries), as measured by the increase in uKIN’s average ranking of genes we aimed to uncover. (B) uKIN is effective in identifying complex disease genes. We demonstrate the versatility of the uKIN framework by integrating OMIM and GWAS data for three complex diseases, ALS, AMD, and epilepsy. For each disease, we compare uKIN’s performance when using OMIM annotated genes as prior information and GWAS hits as new information with α = 0.5, to baseline versions that propagate only information via diffusion from OMIM (left) or GWAS studies (right). In each panel, for each disease, we plot the log2 ratio of the AUPRC obtained by uKIN to that obtained by the baseline method; in all cases, we observe that these values are positive, thereby demonstrating that uKIN outperforms the baseline methods by successfully integrating prior and new information.