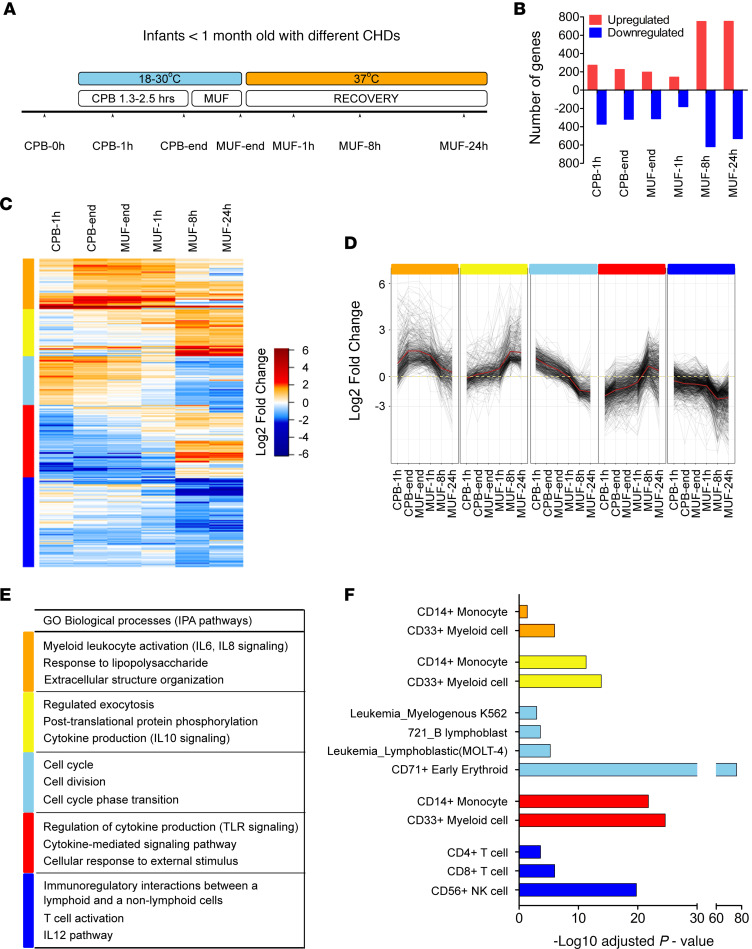
Figure 1. mRNA-Seq analysis of total nucleated cells from pediatric patients undergoing CPB.
(A) The design and time course of the study. Infants with congenital heart diseases (CHDs) underwent CPB surgery that lasted 1.3–2.5 hours on average, followed by a short duration of modified ultrafiltration (MUF). Body temperature was cooled to 18°C–30°C during surgery and quickly rewarmed to 37°C after MUF. Blood samples were collected at 7 time points for mRNA-Seq of total nucleated cells (n = 5 patients). (B) The number of genes that were significantly downregulated or upregulated at different time points compared with the expression level before surgery (CPB-0h) for each patient. Significance was defined as at least 2-fold change with adjusted P < 0.01. (C) Identification of 5 clusters by hierarchical clustering of 2686 differentially expressed genes that were significant at all time points based on Euclidean distances. (D) Kinetics of gene expression within distinct gene expression clusters. Dotted line indicates mean gene expression in each cluster. (E) Pathway enrichment analysis by Metascape and Ingenuity Pathway Analysis (IPA) identified top gene ontology (GO) biological processes being enriched in each cluster. (F) Computational deconvolution of cell subsets in each cluster using GeneAtlas in EnrichR.