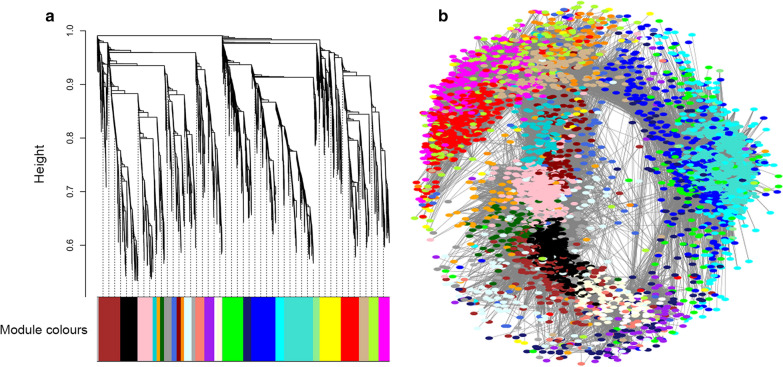
Fig. 2.
An illustration of the identified gene co-expression network modules in T. brucei. a Hierarchical cluster dendrogram. The x-axis represents the co-expression distance of the genes, while the y-axis represents the genes. A dynamic tree cutting algorithm identified the modules by splitting the tree at significant branching points. Modules are represented by different colors as shown by the dendrogram. b Co-expression network from weighted gene co-expression network analysis (WGCNA) based on topological overlap measures (TOMs) > 0.3 for visualization. Each point (or node) on the network represents a gene, and points of the same color form a gene module. Lines (edges) on the network connecting the nodes represent a relationship between the genes