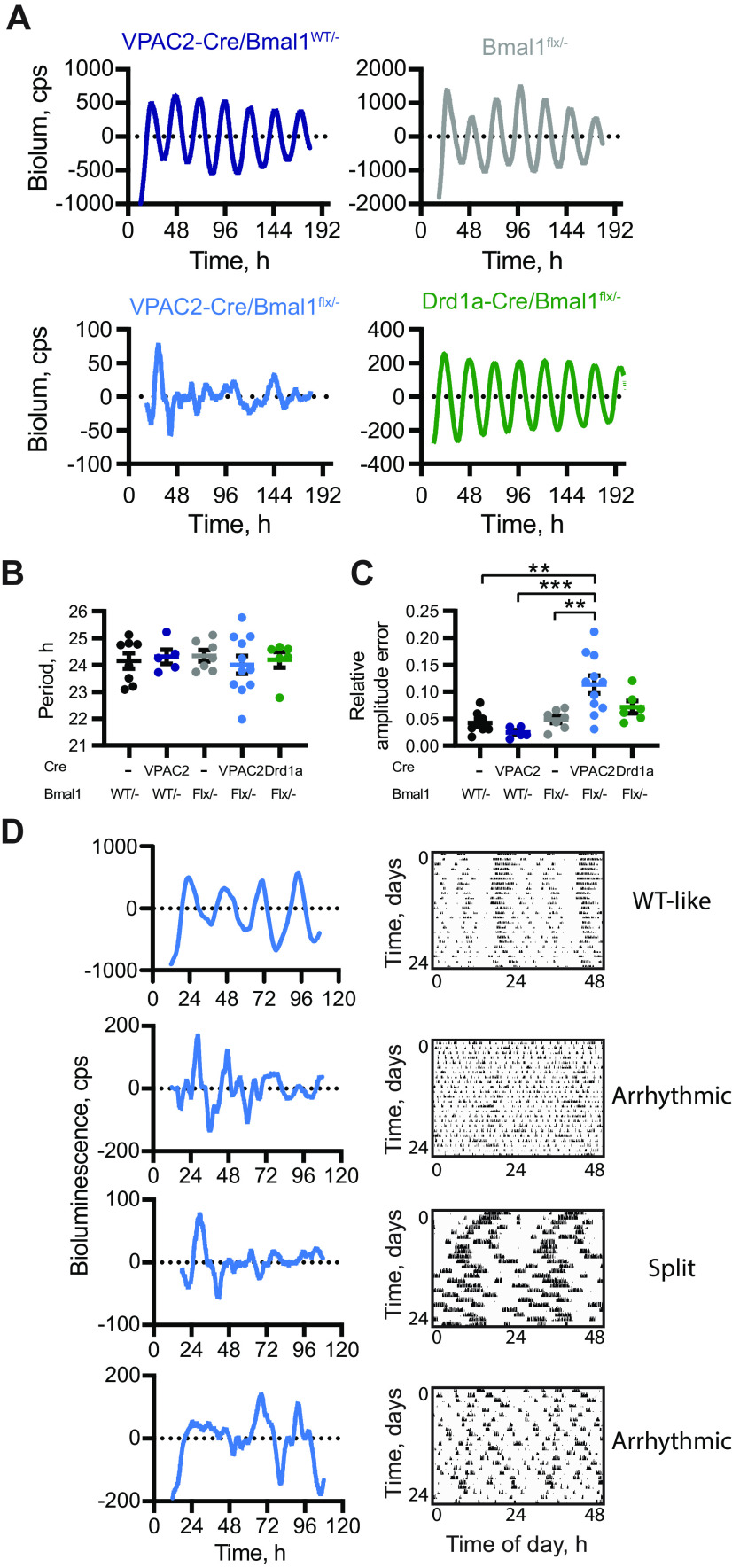
Figure 5.
Deletion of BMAL1 from VPAC2-Cre-expressing cells compromises SCN molecular pace-making. A, Representative, baseline-corrected PER2::LUCIFERASE bioluminescence traces from control, VPAC2-Cre/Bmal1flx/–, and Drd1a-Cre/Bmal1flx/– SCN dissected in dim red light following recording of wheel-running rhythms in DD. B, Period (mean ± SEM) of the first 4 bioluminescent cycles recorded from adult SCN slices; genotypes as in A. n = 8 (Bmal1WT/–), n = 7 (Bmal1flx/–), n = 5 (VPAC2-Cre/Bmal1WT/–), n = 11 (VPAC2-Cre/Bmal1flx/–), and n = 6 (Drd1a-Cre/Bmal1flx/–). C, RAE scores (mean ± SEM) for bioluminescent recordings from SCN slices. n values same as in B. D, Representative PER2::LUCIFERASE bioluminescence rhythms from VPAC2-Cre/Bmal1flx/– SCN slices alongside respective actograms from corresponding mice. “WT-like,” “Arrhythmic,” or “Split” beside the actograms indicates the phenotypic category. **p < 0.01; ***p < 0.001; one-way ANOVA, with Tukey's post hoc test.