Figure 1.
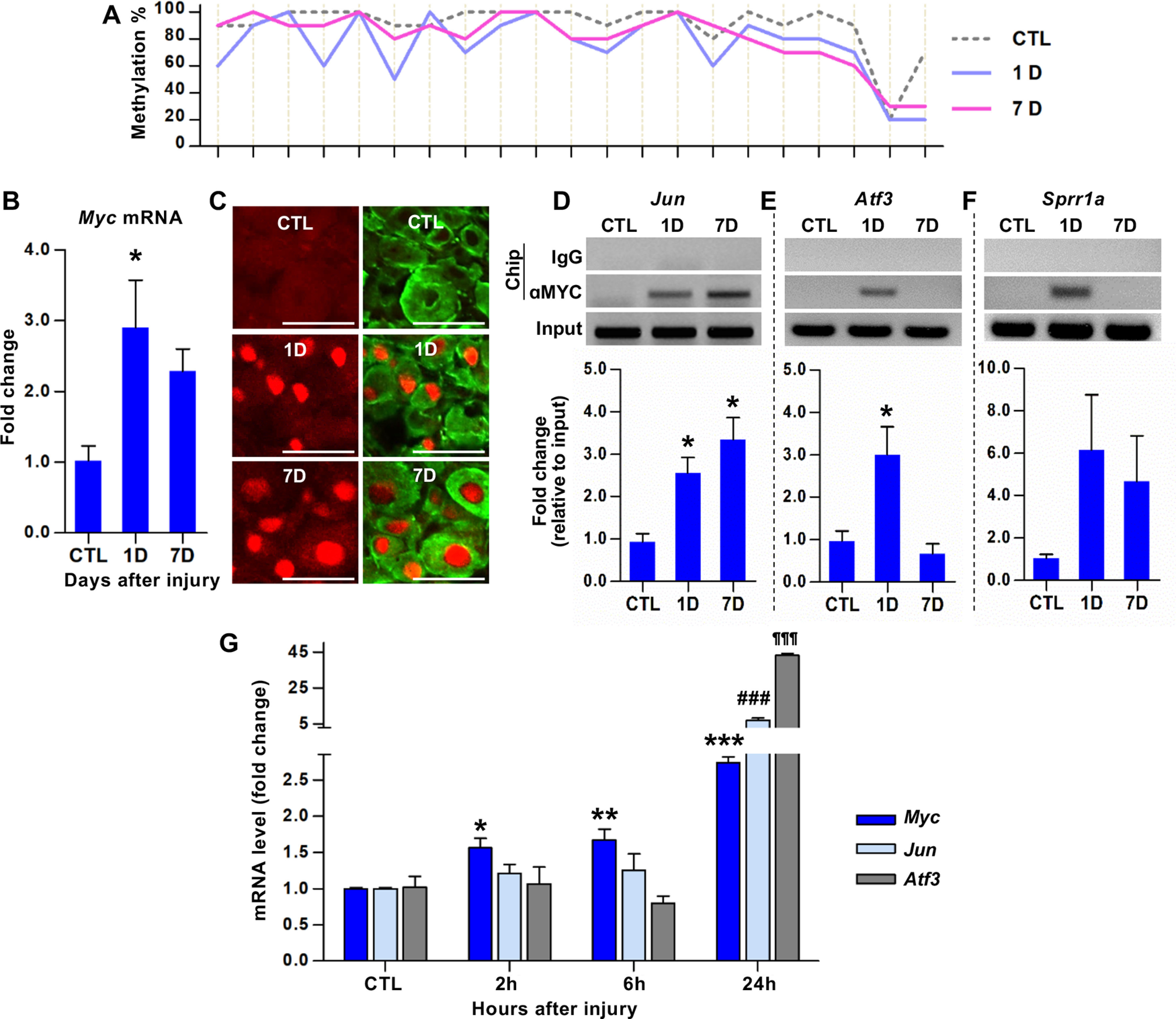
Identification of Myc as a hub transcription factor gene for a subset of the RAGs following preconditioning peripheral nerve injury. A, Bisulfite sequencing analysis of CpGs within the Myc exon 3 sequences using DNA obtained from the DRGs of animals killed at different time points following SNI. The percentage of the clones revealing methylated cytosine was shown on the y axis. Each line crossing the x axis indicates individual CpG sites. B, qRT-PCR results of the Myc gene expression in DRGs following SNI. *p < 0.05 (one-way ANOVA followed by Tukey's post hoc analysis). N = 4 animals per group. C, Representative images of DRG tissue sections immunostained with anti-c-Myc antibodies. Predominantly nuclear c-Myc immunofluorescence signals (red) were observed in DRG neurons visualized by NeuN immunoreactivity (green). Scale bars, 100 μm. D–F, ChIP analysis of c-MYC binding to the promoters of Jun (D), Atf3 (E), and Sprr1a (F). DRG samples were subjected to immunoprecipitation using antibodies against c-MYC, and the immunoprecipitated DNA fragments were amplified by qRT-PCR. *p < 0.05 (one-way ANOVA followed by Tukey's post hoc analysis). N = 3 animals per group. G, qRT-PCR results comparing the expression levels of Myc, Jun, and Atf3 in DRGs at early time points following SNI. For Myc: *p < 0.05; **p < 0.01; ***p < 0.001; compared with control (CTL). For Jun: ###p < 0.001; compared with CTL. For Atf3: ¶¶¶p < 0.001; compared with CTL. Statistical significance was determined by one-way ANOVA followed by Tukey's post hoc analysis. N = 3 animals per time point.
