Figure 3.
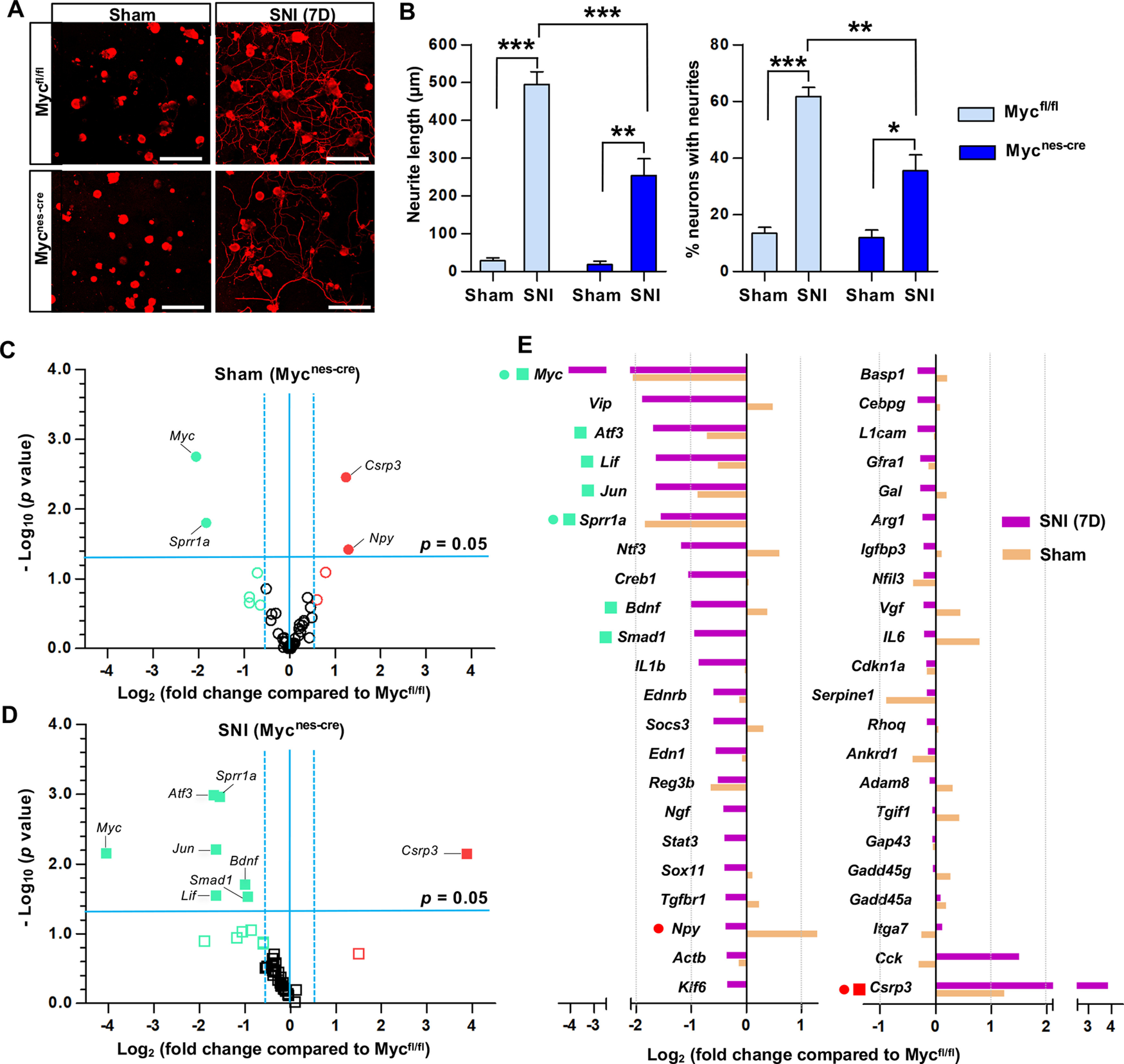
Myc is necessary for the enhancement of regenerative capacity and injury-induced upregulation of a group of RAGs. A, Representative images of cultured mouse DRG neurons. DRGs were obtained from Myc floxed mice (Mycfl/fl) or Myc flox/flox; Nestin-Cre mice (Mycnes-cre), where Myc is deleted in neural lineage cells. Mice with either genotype were sham-operated or underwent SNI, and their L4-L6 DRGs were acutely dissociated 7 d after the surgery and cultured for 15 h before fixation. Neurons and their neurites are visualized by immunoreactivity against βIII tubulin. Scale bars, 50 μm. B, Quantification graphs of the neurite length and the percentage of neurons with neurites longer than 2 times the longest length of cell bodies. *p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA followed by Tukey's post hoc analysis. N = 3 animals per genotype for sham-operated groups, and N = 5 animals per genotype for SNI groups. C, D, Volcano plots of the 44 RAG expressions in DRGs measured using the PCR array comparing the WT (Mycfl/fl) and Myc-deficient (Mycnes-cre) animals either with sham operation (C) or SNI (D). The L4-L6 DRGs were dissected 7 d after sham operation or SNI. Dotted vertical lines indicate a 1.5-fold increase or decrease compared with the PBS control group. Solid horizontal line indicates p = 0.05 (unpaired t test), comparing the WT (Mycfl/fl) and Myc-deficient (Mycnes-cre) animals with either sham operation (C) or SNI (D). Red circles or squares represent the genes revealing a >1.5-fold increase in expression by Myc deletion in the sham operation (C) or SNI (D) condition, respectively. Green circles or squares represent the genes revealing a >1.5-fold decrease in expression by Myc deletion in the sham operation (C) or SNI (D) condition, respectively. Filled circles or squares represent the genes of which changes in expression by Myc deletion were statistically significant (p < 0.05). N = 3 biological replicates per experimental group. E, Quantification graph of gene expression levels of Myc-deficient animals (expressed as fold changes compared with Mycfl/fl) for the 44 RAGs measured using the PCR array. The symbols preceding the gene titles are matched with those designated in the volcano plots (C,D).
