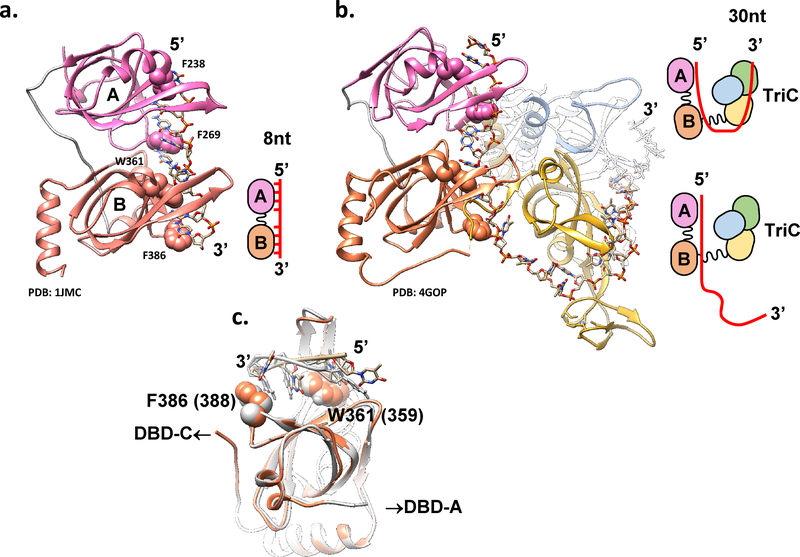
Figure 3: Proposed RPA-ssDNA binding modes.
Proposed structural basis for the distinct RPA-ssDNA binding modes. (a) PDB: 1JMC (76) structure of the human RPA DBD-A (pink) and DBD-B (orange) in complex with 8nt poly(dC) ssDNA represents an 8 nt binding mode schematically depicted on the right. Atoms of the four conserved aromatic residues F238 and F269 (in DBD-A) and W361 and F386 (in DBD-B) are shown as spheres. (b) PBD: 4GOP (50), which represents the high affinity (30 nt) ssDNA binding mode of Ustilago maydis RPA is shown with the same orientation of the DBDs A and B. Cartoon on the right represents the proposed transition between the 8nt and 30nt binding modes. (c) Overlap of the DBD-B from 4GOP (orange) and 1JMC (light grey) suggests different paths the ssDNA can take through this DBD likely due to the presence of the flexible linker connecting DBD-B and DBD-C. See text and (50) for extended discussion.