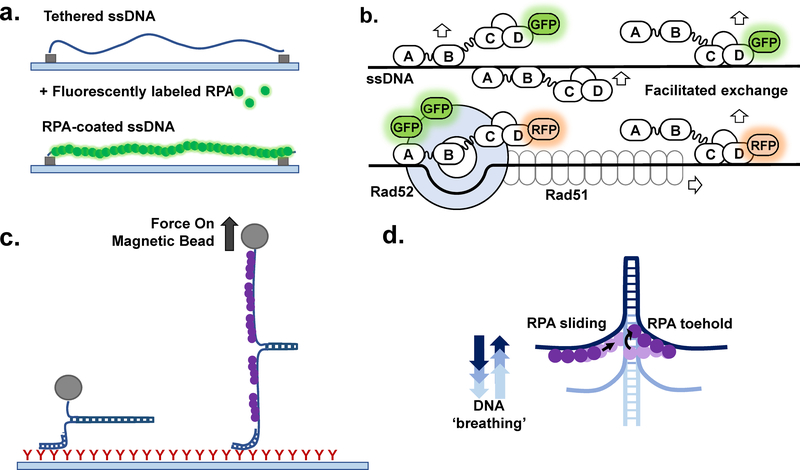
Figure 4: Single-molecule interrogation of the RPA-ssDNA complex.
(a) Schematic representation of an ssDNA curtain experiment (see (11,39,96) for details), side view. The ssDNA molecules are stretched parallel to each other over the surface of the TIRFM slide. Binding of the RPA-GFP (green) is visualized as appearance of the fluorescence along the DNA molecules. (b) The DNA curtains experiments allowed to propose the mechanism underlying facilitated exchange of RPA on ssDNA. When no additional RPA is present in the solution, RPA molecules remain stably bound to ssDNA even though their individual binding modules may microscopically dissociate from and rebind to ssDNA (e.g. transition between 8 nt and 30 nt binding modes). In the presence of unlabeled RPA in solution, microscopic dissociation of the trimerization core (30 nt → 8 nt transition) opens a landing spot for the RPA from solution and subsequent exchange of the GFP labeled protein on the ssDNA with the unlabeled counterpart. Two color experiments with RFP-labeled RPA and GFP-labeled Rad52 showed that the recombination mediator Rad52 stabilizes some RPA molecules on ssDNA resulting in the RPA-Rad52 clusters from which the Rad51 nucleoprotein filament grows. (c) Magnetic tweezer experiment to characterize the RPA-mediated DNA duplex melting (47). (d) These experiments suggested that a microscopic association of the individual RPA DBDs creates a “toehold” that traps spontaneously melted DNA duplex at the ssDNA-dsDNA junction and promotes duplex destabilization by RPA.