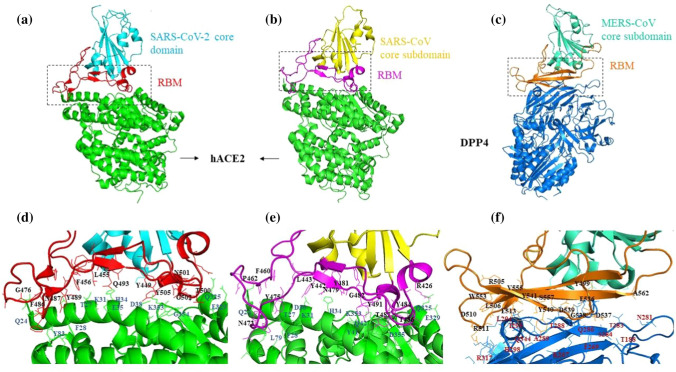
Fig. 2.
Crystal structures of human betacoronavirus S1 receptor binding domains in complex with their receptor. a) Structure of the SARS-CoV-2 receptor-binding domain (RBD) showing the core domain (cyan) and the receptor-binding motif (RBM, red) complexed with human ACE2 (green; PDB ID: 6VW1). b) Structure of the SARS-CoV RBD showing the core domain (yellow) and RBM (pink) complexed with human ACE2 (green; PDB ID: 2AJF). c) Structure of the MERS-CoV RBD showing the core domain (sea green) and RBM (orange) complexed with human DPP4 (blue; PDB ID: 4L72). d) Interface between the SARS-CoV-2 RBM and ACE2. Critical residues of RBM and ACE2 involved in the interaction are labelled in black and blue, respectively. e) Interface between the SARS-CoV RBM and ACE2. Critical residues of RBM and ACE2 involved in the interaction are labelled in black and blue, respectively. f) Interface between the MERS-CoV RBM and DPP4. Interacting residues of RBM and DPP4 are labelled in black and red, respectively. The crystal structures with their respective PDB IDs were downloaded from the RCSB Protein Data Bank (https://www.rcsb.org/), and the figures were prepared using PyMol (Version 2.0 Schrodinger, LLC).