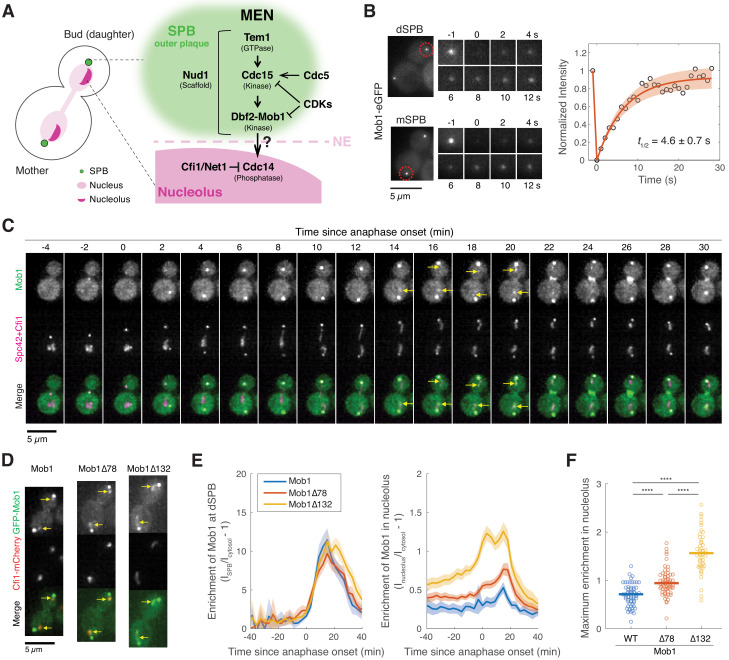
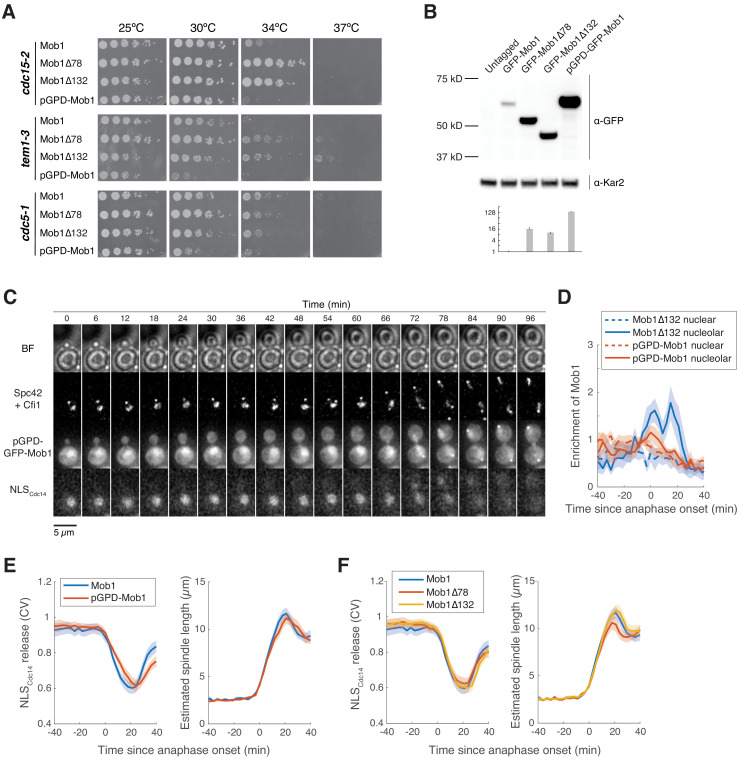
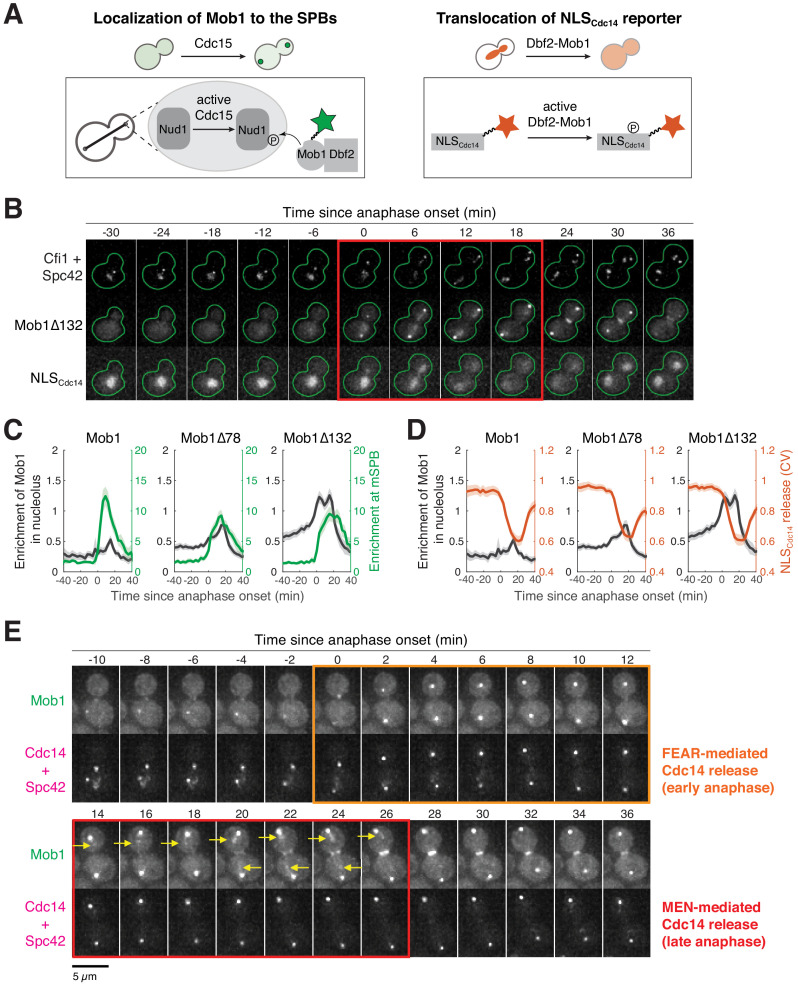
Figure 1. Dbf2-Mob1 transiently localizes to the nucleolus in late anaphase.
(A) Major components of the mitotic exit network (MEN) and their subcellular localization. (B) Fluorescence recovery after photobleaching (FRAP) analysis of Mob1-eGFP (A39695). Red circles indicate the area of photo-bleaching. Cells were grown and imaged at room temperature in SC medium + 2% glucose. Graph to the right represents average measurements of double normalized fluorescence intensities (n = 6 cells) after correcting for photo-bleaching during acquisition. Red curve is the average fit and shaded area represents standard deviation (SD) of the fits. Half recovery time ± SD is indicated. (C) Localization of Mob1 during the cell cycle. A40257 (with Mob1-eGFP, Cfi1-mCherry and Spc42-mCherry) cells were grown at room temperature in SC medium + 2% glucose and imaged every minute for 2 hr. Arrows highlight the nucleolar localization. (D) Nucleolar localization of full-length (A39931) and N-terminally truncated (A39933 and A39935) Mob1. Cells were grown at room temperature in SC medium + 2% glucose and imaged every 3 min for 4 hr. Arrows highlight the nucleolar localization. (E) Enrichment of Mob1 (A41211, n = 62 cells), Mob1Δ78 (A41212, n = 60 cells), and Mob1Δ132 (A41213, n = 48 cells) at the daughter spindle pole body (dSPB; left) and in the nucleolus (right) as a function of cell cycle progression. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. Single cell traces were aligned based on anaphase onset, as defined as spindle length >3 μm (Figure 1—figure supplement 1F, measured based on SPB marker Spc42-mCherry), and averaged. Solid lines represent the average, and shaded areas represent 95% confidence intervals. (F) Maximum enrichment of full-length Mob1 (WT) and truncated Mob1 (Mob1Δ78 and Mob1Δ132) in the nucleolus in anaphase of cells in (E). Solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test.