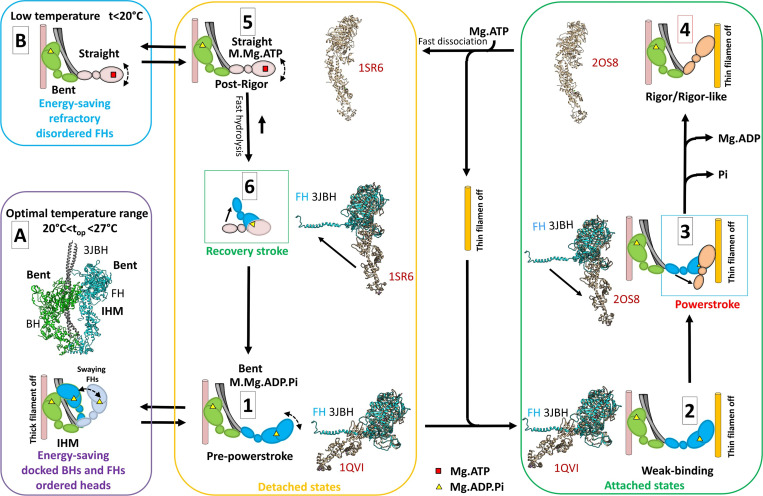
Figure 7.
Two ATP energy-saving mechanisms proposed in tarantula skeletal muscle. A cross-bridge cycle for tarantula skeletal muscle, based on previous work by Robert-Paganin et al. (2020), is shown and includes actin-detached steps (yellow box) and actin-attached steps (green box). (A) The energy-saving IHM (PDB accession no. 3JBH) allows quick incorporation of the detached, swaying FHs to the cycle (Padrón et al., 2020). The FHs, which are in the bent, prepower stroke structure (similar to PDB accession no. 1QVI, shown in tan in steps 1 and 2 and in Fig. 1 B) containing Mg.ADP.Pi (Zoghbi et al., 2004), can sway away and back by Brownian motion (Brito et al., 2011; step 1) and weakly attach to the switched-off thin filaments (step 2). When the thin filament is switched on by the tropomyosin/troponin Ca2+ switch (Craig and Lehman, 2001), the FHs attach strongly, going through the power stroke and producing force and displacement (step 3) transitioning, after Pi and then ADP release, to a nucleotide-free rigor/rigor-like structure similar to PDB accession no. 2OS8 (step 4). The binding of Mg.ATP to the rigor/rigor-like FHs induces fast dissociation of the heads from actin (step 5), followed by a rapid hydrolysis of Mg.ATP to Mg.ADP.Pi during the recovery stroke (step 6), reproducing the prepower stroke (step 1). We propose that there are two different mechanisms for ATP energy saving in the relaxed, detached states involving (1) IHM energy saving by helically ordered docked BHs and FHs, with swaying FHs enabled to quickly produce force as they are all in the bent prepower stroke Mg.ADP.Pi-containing state (A, purple box); and (2) the refractory, energy-saving, disordered, and functionally disabled heads that need to be reprimed during the recovery stroke to achieve the prepower stroke, enabling them to produce force (B, blue box). The structures in steps 1–3 and 6 show the superimposition of the motor domains of the FH of PDB 3JBH in the same orientation (without the regulatory and essential light chains) with the crystal structures PDB 1QVI, PDB 2SO8, and PDB 1SR6.