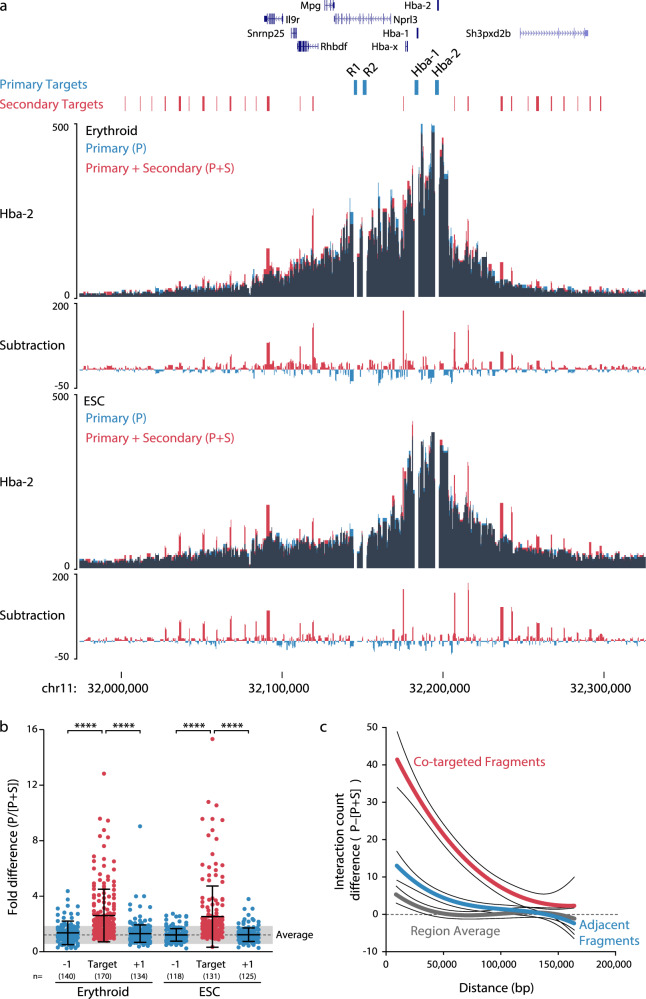
Fig. 2. Co-targeting results in variable-magnitude, target-specific bias.
a 3C libraries from mouse erythroid (n = 3 independent experiments) and embryonic stem cells (ESC; n = 3 independent experiments) were captured with either a pool of probes containing eight primary (P) viewpoints, or a pool of probes containing both the primary viewpoints and 54 additional, or secondary (S), viewpoints. Captured fragments were analyzed only for the primary viewpoints. Data is shown as an overlay for the Hba-2 capture viewpoint, with dark areas showing where signal overlaps. b Comparison of the relative difference in interaction counts at co-targeted fragments and the adjacent fragments (±1). Each dot (n shown) represents the average skew after capture in three independent 3C libraries from each of seven primary viewpoints. The difference in n between Erythroid and ESC arises from a poor mapability of the beta-globin locus in this ESC cell line. Average with one standard deviation (gray shading) is shown for all fragments within 160 kb of the primary targets. c Distance dependent difference in signal caused by co-targeting compared with adjacent fragments and the region average. ****p < 0.0001 using a two-sided Mann–Whitney U-test. Bars show mean and one standard deviation. Source data are available in the Source Data file.