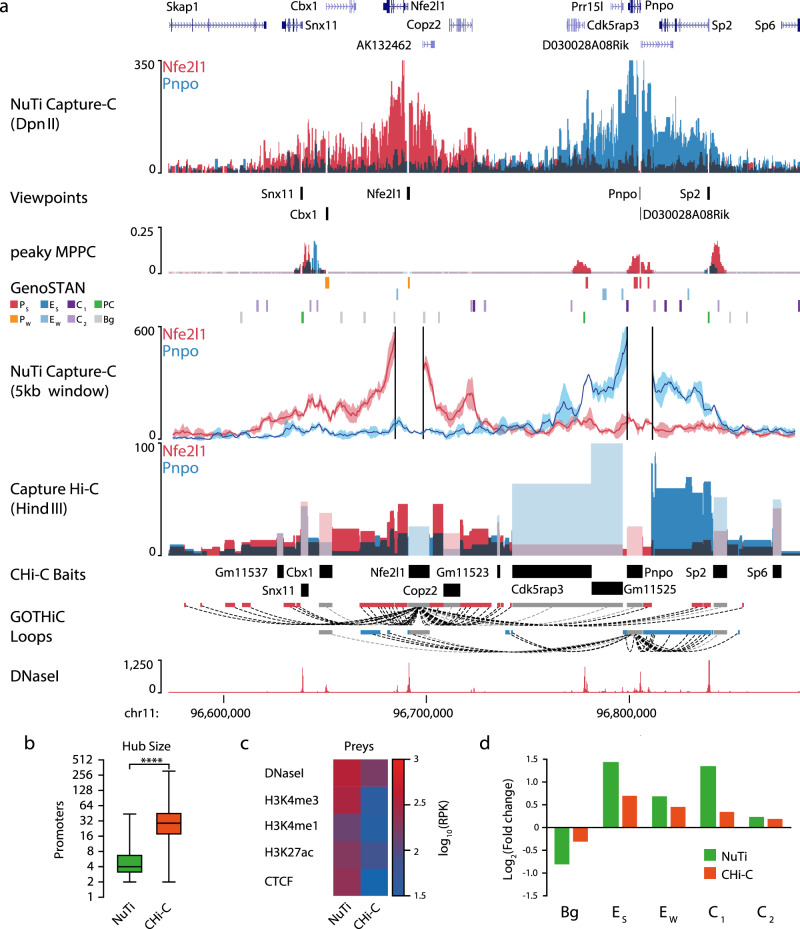
Fig. 4. Comparison of capture resolution at the Nfe2l1 and Pnpo promoters.
a Sequence tracks showing the difference between high-resolution 3C (DpnII, NuTi Capture-C) and low-resolution 3C (HindIII, Capture Hi-C) from nearby gene promoters (mm9, chr11:96,572,876-96,883,917) in erythroid cells. Tracks in order: UCSC gene annotation, cis-normalized mean interactions per DpnII fragment using NuTi Capture-C (n = 3 independent 3C libraries), NuTi Capture-C viewpoints, peaky Marginal Posterior Probability of Contact (MPPC) scores with fragments with MPPC ≥ 0.01 darker, GenoSTAN open-chromatin classification, windowed mean interactions using NuTi Capture-C, total supporting reads per HindIII fragment with CHi-C (n = 2; co-targeted fragments are lighter in color), CHi-C bait fragments, loops between reported significantly interacting fragments (co-targeting loops are colored gray), erythroid tracks for open chromatin (DNaseI). Note overlapping blue and red signals appear darker in color (NuTi Capture-C, peaky MPPC, CHi-C). b Number of interacting promoters identified as present in promoter-hubs. ****p < 0.0001 a two-sided Mann–Whitney U-test. For NuTi n = 4339 promoter viewpoints, and for CHi-C n = 19,683 promoter viewpoints. Box and Whiskers show: minima 25th percentile, median, 75th percentile and maxima. c Average chromatin signature in mouse erythroid cells over fragments identified as being significantly interacting with promoters by NuTi Capture-C (Nu-3C) and Capture Hi-C (CHi-C). RPK: Reads per kilobase. d Enrichment of different classes of open-chromatin element in fragments identified as being significantly interacting with active promoters. ES: Enhancer (Strong H3K27ac), EW: Enhancer (Weak H3K27ac), C1: CTCF near promoter/enhancer, C2: CTCF, Bg: Background. Source data are available in the Source Data file.