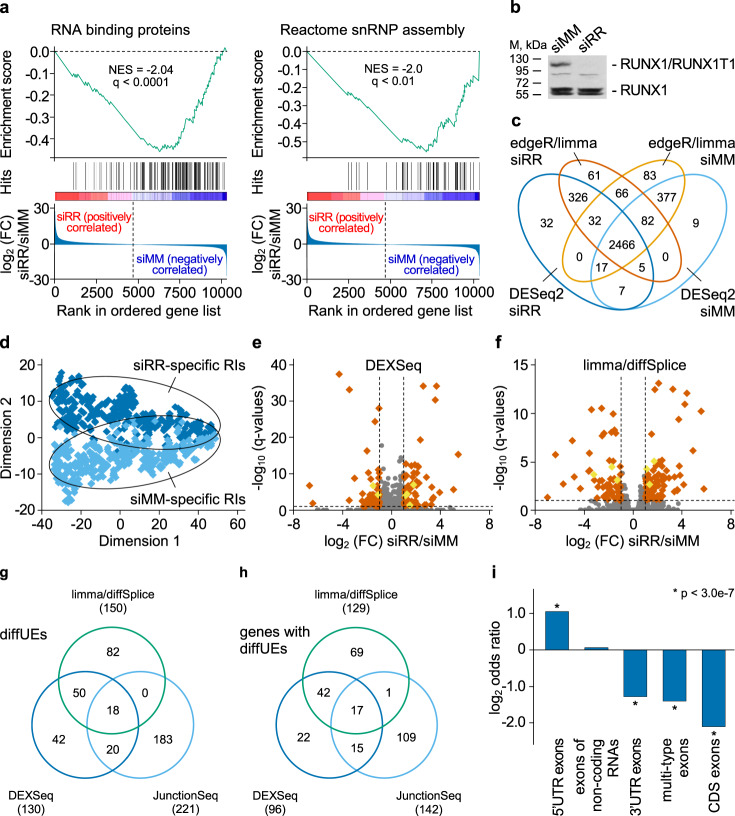
Fig. 1. Knockdown of RUNX1/RUNX1T1 affects exon usage in Kasumi-1 cells.
a Gene set enrichment analysis plots for RNA binding proteins55 and the Reactome snRNP assembly pathway. NES, normalized enrichment score; q, false discovery rate. b Western blot of RUNX1 and RUNX1/RUNX1T1 protein levels in Kasumi-1 cells following RUNX1/RUNX1T1 knockdown. A representative result from one of the three experiments is shown. siMM, mismatch siRNA; siRR, RUNX1/RUNX1T1 siRNA. c Distribution of retained introns (RIs) between the transcriptomes of the siRR-treated and siMM-treated Kasumi-1 cells. The RIs were detected using the DESeq2 and edgeR/limma algorithms. d Scatter plot showing the distribution of siRR-specific and siMM-specific RIs after multidimensional reduction of RIs expression data using t-distributed stochastic neighbor embedding. The RIs were detected using the edgeR/limma algorithm, and very similar results were observed with DESeq2. e, f Vulcano plots showing differential usage of exons identified by DEXSeq e or limma/diffSplice f algorithms. Orange and yellow rhombi indicated differentially used canonical exons or RIs, respectively, with more than 2-fold change and q < 0.1. g Venn diagram showing overlap between differentially used exons (diffUEs) identified by DEXSeq, limma/diffSplice and JunctionSeq algorithms. h Venn diagram showing overlap of genes with diffUEs identified by the DEXSeq, limma/diffSplice and JunctionSeq algorithms. i Classification of diffUEs according to the functional type of exons. Genomic coordinates of the reference exons were extracted from Ensembl models of the human genes. diffUEs were intersected with reference exons and the overlaps counted. Fisher’s two-sided exact test was performed to detect over-represented or under-represented exon types among the diffUEs against non-differential exons (non-diffUEs).