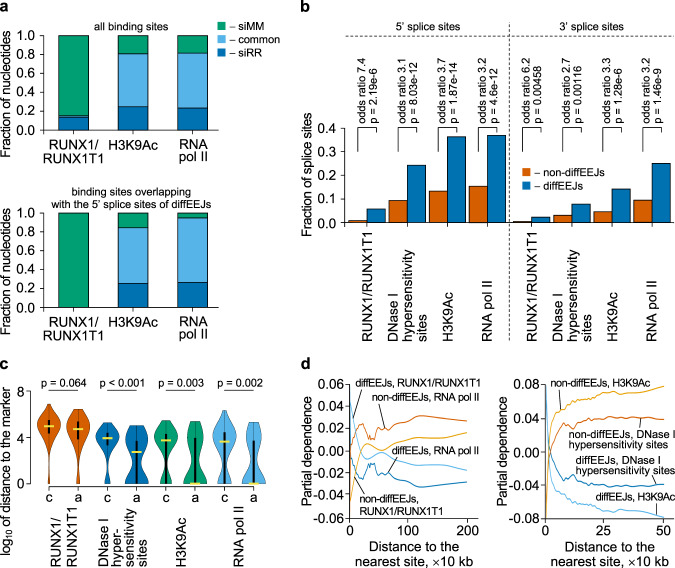
Fig. 4. RUNX1/RUNX1T1-dependent differential splicing events in Kasumi-1 cells are associated with open chromatin.
a Column graph showing the distribution of RUNX1/RUNX1T1, H3K9Ac and RNA pol II occupied sites with or without RUNX1/RUNX1T1 knockdown. The top panel shows the distribution for all sites, the bottom panel only shows distribution of binding sites overlapping with the 5’ splice sites of diffEEJs. b Association of diffEEJs splice sites with open chromatin. P-values were calculated with Fisher’s two-sided exact test. c Violin graph depicting the positional relationships between canonical (c; n = 117) and alternative (a; n = 57) 5’ splice sites of diffEEJs and marks of open chromatin. P-values were calculated with two-sided Mann–Whitney U test. Yellow horizontal thick lines represent the median of distances distribution, the black vertical rectangular boxes show the interquartile range, and the black vertical lines are the 95% confidential interval. d Effect of the proximity to RUNX1/RUNX1T1 binding sites, RNA pol II peaks, H3K9Ac histone modifications and DNase I hypersensitivity sites on the probability of the diffEEJs and non-diffEEJs in the transcriptome of leukemia cells.