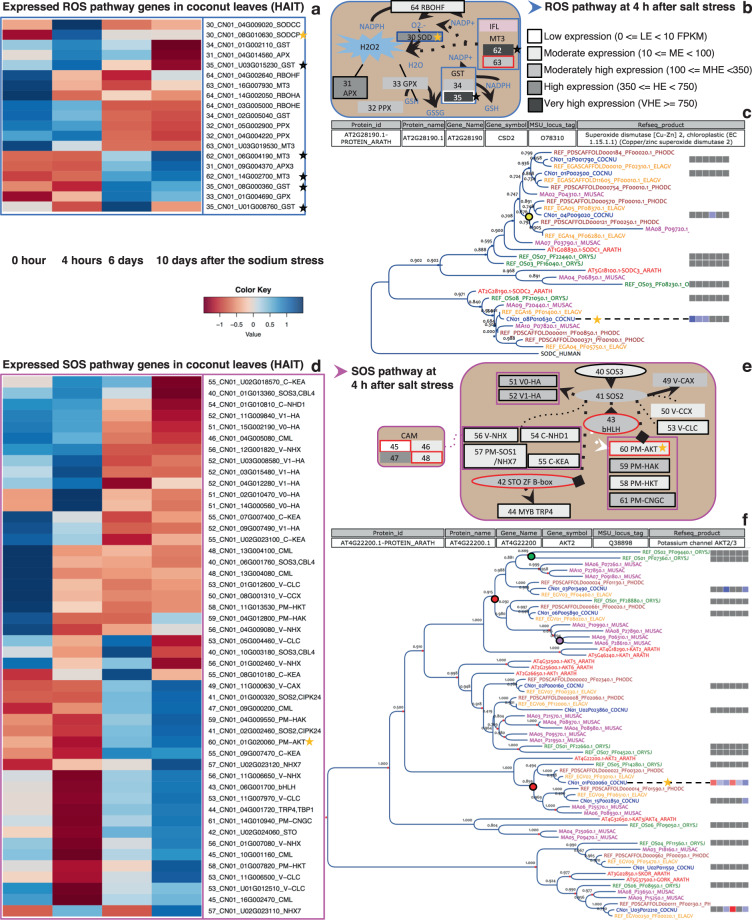
Fig. 6. Early gene regulation during salt stress in coconut leaves.
a Normalized transcriptional expression level in Hainan tall leaves at time points 0, 4 h, 6 days, and 10 days after salt stress for the DE and CHE genes of the ROS pathway. Each gene is identified by its expression class number (ECN), gene identifier, and function symbol. b Schematic representation of the ROS signaling pathway at 4 h (see Supplementary Fig. S9 for more details). c Gene phylogeny of the SODC family across coconut (COCNU), date palm (PHODC), oil palm (ELAGA), banana (MUSAC), japonica rice (ORIZJ), and Arabidopsis (ARATH). Colored bullets represent WGDs (yellow, red, purple, and green for respectively p, τ, α/β/γ, and ρ/σ WGDs). Expression differential of coconut genes is displayed with blue and red squares for up and downregulation, respectively. The first three columns are for the Hainan tall variety 4 h, 6 days, and 10 days) and the last three for the aromatic dwarf variety. Note that SODCP candidate gene CN01_08G010630 (yellow star) is orthologous to AT2G28190 whose annotation is given in the box above the tree. See Supplementary Note 1 for results on the MT3 and GST genes (gray stars). d, e Same as (a, b) for the SOS pathway. f Same as (c) for the Shaker gene family. Note that AKT2 candidate gene CN01_01G020060 (yellow star) is orthologous to AT4G22200 whose annotation is given in the box above the tree.