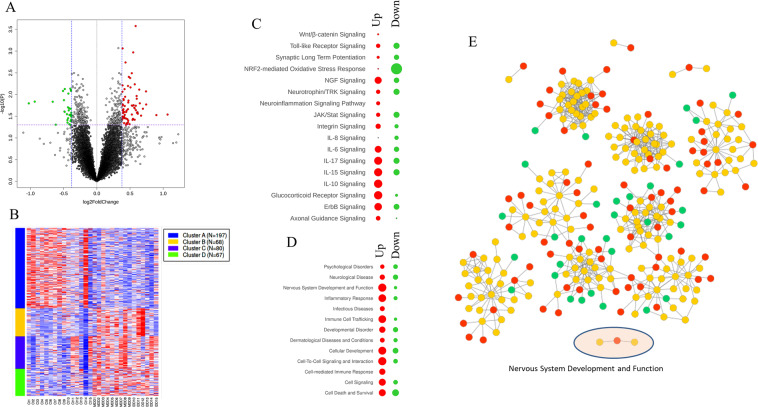
Fig. 1. Volcano plot and heatmap based on total fraction RNA-seq data.
A Volcano plot was created by all differentially expressed genes (14,005 genes). Y-axis shows the mean expression value of log 10 (p value), and the x-axis displays the log 2-fold change value. Blue vertical line means 1.3-fold change [log 2 (0.584)]. The purple horizontal line means p = 0.05 [−log 10 (1.30)]. B k-Means clustering was done over heatmap of only significant (p < 0.05) genes made over standard deviation normalization (212 upregulated and 199 downregulated genes). Canonical pathway, disease and function, and gene–gene networks were analyzed using 73 upregulated and 31 downregulated genes (>1.3-fold). The significant results of the canonical pathway (C) and disease and function (D) are shown in red (up) and green (down) circles. Spots/circles size is the function of −log(base = 10) of Fisher’s exact test enrichment p value. E IPA gene–gene network yielded 13 disease function modules. All 13 subnetwork were merged to build one network. Red nodes represent upregulated genes, whereas green nodes represent downregulated genes. Other genes are presented in orange color. Ct control, MDD major depressive disorder.