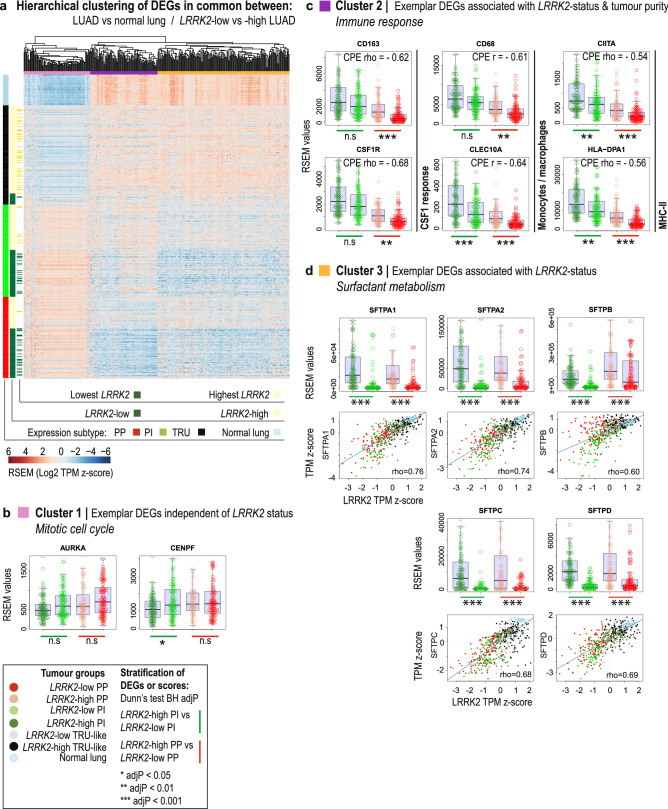
Figure 2.
The transcriptional landscape of LRRK2 repression in lung adenocarcinoma patients. (A) Heatmap depicting the hierarchical agglomerative clustering of genes differentially expressed in common between two biological contexts in TCGA LUAD tumors, each with a marked reduction of LRRK2 expression: (1) across all tumors versus normal lung and (2) in the lowest LRRK2 expressing tumors versus the highest LRRK2 expressing tumors. Columns: n = 385 DEGs with absolute fold change of median RSEM value > 2; Rows: n = 59 normal lung and n = 517 LUAD tumors, ordered by tissue, LRRK2 expression status and expression subtype. (B) Exemplar DEGs identified in Cluster 1, enriched for genes that act in mitotic cell cycle (Metascape algorithm; q < 0.05), stratified by tumor group (multicolour boxplots of exemplar gene expression; Dunn’s test BH adjP > 0.05). Tumor groups represent the combined sample status for LRRK2 expression and non-TRU subtype. (C) Exemplar DEGs identified in Cluster 2, associated with tumor purity (consensus purity estimate or CPE; Spearman’s correlation coefficient ≥ 0.5 with Holm’s corrected P < 0.0001) and enriched for immune response genes (Metascape algorithm; q < 0.05), stratified by tumor group (multicolour boxplots of exemplar gene expression; Dunn’s test BH adjP < 0.01). (D) Exemplar DEGs identified in Cluster 3, enriched for genes that act in surfactant metabolism (Metascape algorithm; q < 0.05), stratified by tumor group (multicolour boxplots of exemplar gene expression; Dunn’s test BH adjP < 0.001) or plotted against LRRK2 expression (multicolour scatterplots; Spearman’s correlation coefficient ≥ 0.6 with Holm’s corrected P < 0.0001; standardized RSEM Transcripts Per Million or TPM).