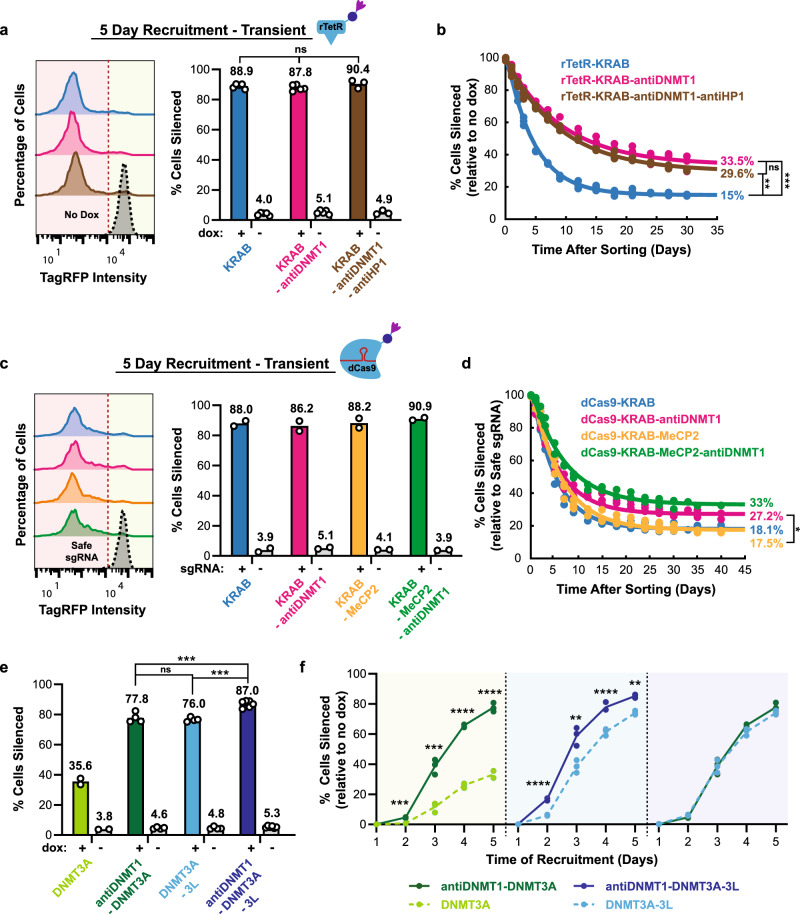
Fig. 3. Nanobody-mediated enhancement of KRAB and DNMT3A repression.
a Left: fluorescence distributions of the reporter gene after transient expression of rTetR-effector fusions and recruitment by dox treatment for 5 days, measured using flow cytometry. Right: the means of percent cells silent (to the left of the red dotted line in the left-side plots) are shown as bars. Each dot represents an independently transfected biological replicate (KRAB: n = 5; KRAB-antiDNMT1: n = 5; KRAB-antiDNMT1-antiHP1: n = 3; statistical analysis by one-way ANOVA). b After recruitment with rTetR-effector fusions for 5 days, silenced cells were sorted and memory dynamics were measured by flow cytometry throughout 30 days. Data represent three biological replicates and are fitted with an exponential decay curve (“Methods”). Two-tailed Tukey’s test analysis at day 30 after sorting (KRAB vs. KRAB-antiDNMT1: ***p = 0.00022; KRAB vs. KRAB-antiDNMT1-antiHP1: **p = 0.0016). c Fluorescence distributions after transient expression and targeting of dCas9-effector fusions to the TetO sites upstream of the reporter gene (+) or to a safe-targeting control site (−) for 5 days. Means are from two biological replicates. d After targeting the dCas9-effector fusions for 5 days, silenced cells were sorted, and memory dynamics was measured by flow cytometry throughout 40 days. Each dot is a biological replicate (KRAB: n = 2; KRAB-antiDNMT1: n = 3; KRAB-MeCP2: n = 3; KRAB-MeCP2-antiDNMT1: n = 2) and are fitted with an exponential decay curve (“Methods”). Two-tailed Tukey’s test at day 40 after sorting (KRAB-MeCP2 vs. KRAB-antiDNMT1: *p = 0.040). e Transient expression and recruitment of rTetR-based DNA methyltransferase combinations to the reporter gene for 5 days (DNMT3A: n = 2; antiDNMT1-DNMT3A: n = 4; DNMT3A-3L: n = 4; antiDNMT1-DNMT3A-3L: n = 6). Statistical analysis by two-tailed Tukey’s test (antiDNMT1-DNMT3A vs. antiDNMT1-DNMT3A-3L: ***p = 0.00044; DNMT3A-3L vs. antiDNMT1-DNMT3A-3L: ***p = 0.00015). f Corresponding silencing dynamics of rTetR-based combinations in e throughout 5 days of recruitment (n = 3 replicates). Statistical analysis by two-tailed Tukey’s test (DNMT3A vs. antiDNMT1-DNMT3A: ***p2 = 0.00023, ***p3 = 0.00051, ****p4 = 4.3e − 8, ****p5 = 3.5e − 7 | DNMT3A-3L vs. antiDNMT1-DNMT3A-3L: ****p2 = 5.3e − 8, **p3 = 0.0033, ****p4 = 5.1e − 5, **p5 = 0.0019). Source data are available in the Source Data file.