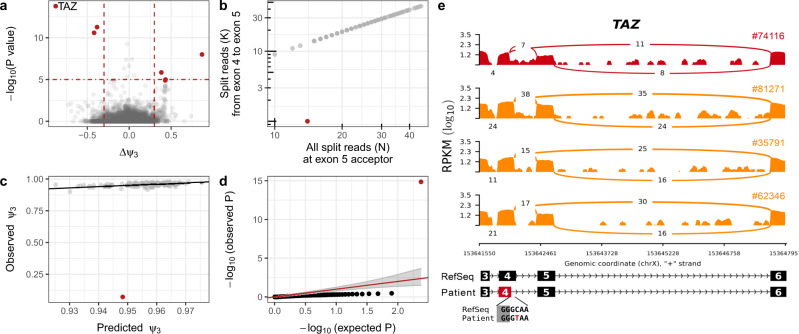
Fig. 7. Detection of a pathogenic splicing defect using FRASER.
a Gene-level significance (−log10(P), y-axis) versus effect (observed minus expected ψ3, x-axis) for alternative donor usage for individual 74116. Six genes (red dots, including TAZ) passed both the genome-wide significance cutoff (horizontal dotted line) and the effect size cutoff (vertical dotted lines). b Number of split reads spanning from the fourth to fifth exon (y-axis) against the total number of split reads at the acceptor site of the fifth exon (x-axis) of the TAZ gene. Sample 74116 (red) deviates from the cohort trend. c Observed (y-axis) against FRASER-predicted (x-axis) ψ3 values for the data shown in b. d Quantile-quantile plot of observed P values (−log10(P), y-axis) against expected P values (−log10(P), x-axis) and 95% confidence band (gray) for the data shown in b. Sample 74116 (red) shows an unexpectedly low P value. e Sashimi plot of the exon-truncation event in RNA-seq samples of the TAZ-affected (red) and three representative TAZ-unaffected (orange) individuals. The RNA-seq read coverage is given as the log10 RPKM-value (Reads Per Kilobase of transcript per Million mapped reads, y-axis) and the number of split reads spanning an intron is indicated on the exon-connecting line. e (bottom) the gene model of the RefSeq annotation is depicted in black and the aberrantly spliced exon is colored in red. The insert depicts the donor site-creating variant of the affected individual 74116. a, d P values were calculated two-sided with the beta-binomial distribution, and significance was determined based on FDR after adjusting for multiple comparisons (“Methods” section).