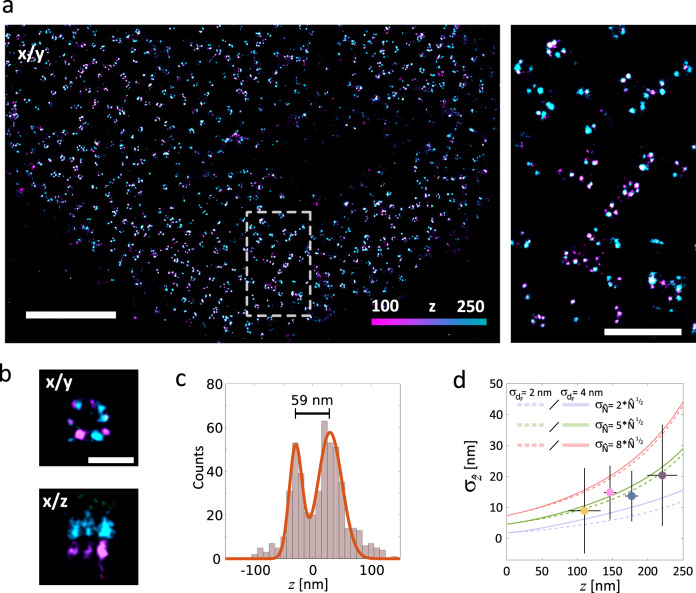
Fig. 4. NUP107 immunolabeled for dSTORM and super-resolved in 3D using SIMPLER.
a Left: top view of dSTORM image of NUP107-mEGFP in HeLa Kyoto cells, labelled with primary anti-GFP antibody conjugated to Alexa Fluor 647. Right: magnified view of the region marked. Four independent experiments were performed with similar results. b Average nuclear pore complex showing the archetypal eightfold symmetry (xy top-view, top) and the organization in nuclear and cytoplasmic rings (xz side-view, bottom) reconstructed by SIMPLER (n = 4). Top- and side-views have the same z-colour scale as defined in a. c Histogram of z-positions of the nuclear pore complex shown in b yields 59-nm separation between the nuclear and cytoplasmic rings. d Median values of different z-positions obtained experimentally from 161 dSTORM single-molecule traces overlapped with theoretical curves of σz for 0 = 10,000 and = 2x, 5x or 8x for σdF = 2 and 4 nm. Error bars represent the standard deviation of and z. The number of single-molecule traces analysed in each interval was 59 (for z = 35–130 nm); 45 (for z = 130–160 nm); 30 (for z = 160–190 nm) and 23 (for z = 190–250 nm). Data include localizations from four biologically independent samples (four HeLa Kyoto cells from independently prepared samples). Scale bars represent 2 µm (a, left); 500 nm (b) and 100 nm (c). Number of localizations kept after frame-filtering step: b 1101 (out of 2889).