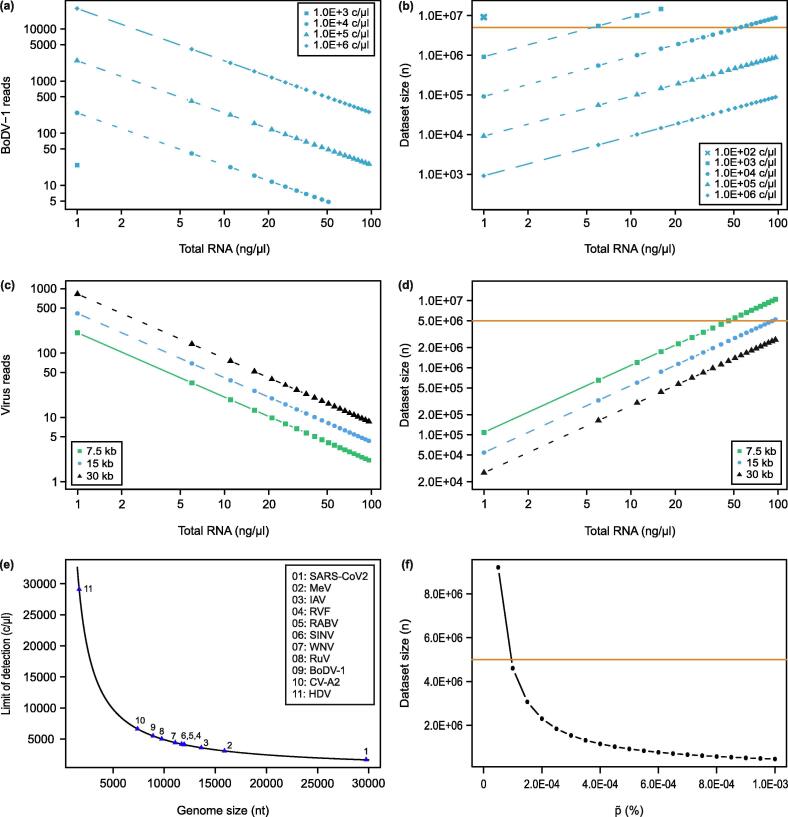
Fig. 2.
Theoretical evaluation of impact factors for mNGS analytical sensitivity. (a) Amount of BoDV-1 reads in relation to the total RNA- and virus-concentration at dataset size 5.0E + 06 reads. No BoDV-1 reads were calculated for 1.0E + 02 c/µl. (b) Minimal dataset size (n) required to detect at least one BoDV-1 read depending on the RNA- and virus-concentration. Threshold (orange line) was set at 5.0E + 06 read dataset size. (c) Amount of virus reads depending on the genome length and RNA-concentration. Virus concentration at 1.0E + 04 c/µl and data set size at 5.0E + 06 reads. (d) Minimal dataset size (n) required to detect at least one virus read depending on the genome length at a virus-concentration of 1.0E + 04 c/µl. Threshold (orange line) was set at 5.0E + 06 read dataset size. (e) Limit of detection of mNGS depending on the genome length of RNA viruses. Dataset size at 5.0E + 06 reads and RNA-concentration at 30 ng/µl. Severe acute respiratory syndrome coronavirus 2, SARS-CoV-2; measles virus, MeV; Influenza A virus, IAV; Rift Valley fever virus, RVFV; Rabies lyssavirus, RABV; Sindbis virus, SINV; West Nile virus, WNV; Rubella virus, RuV; Borna disease virus 1, BoDV-1; Coxsackievirus A, CV-A2; Hepatitis delta virus, HDV. (f) Minimal dataset size (n) required to detect at least one pathogen read depending on . Threshold (orange line) was set at 5.0E + 06 read data depth. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)