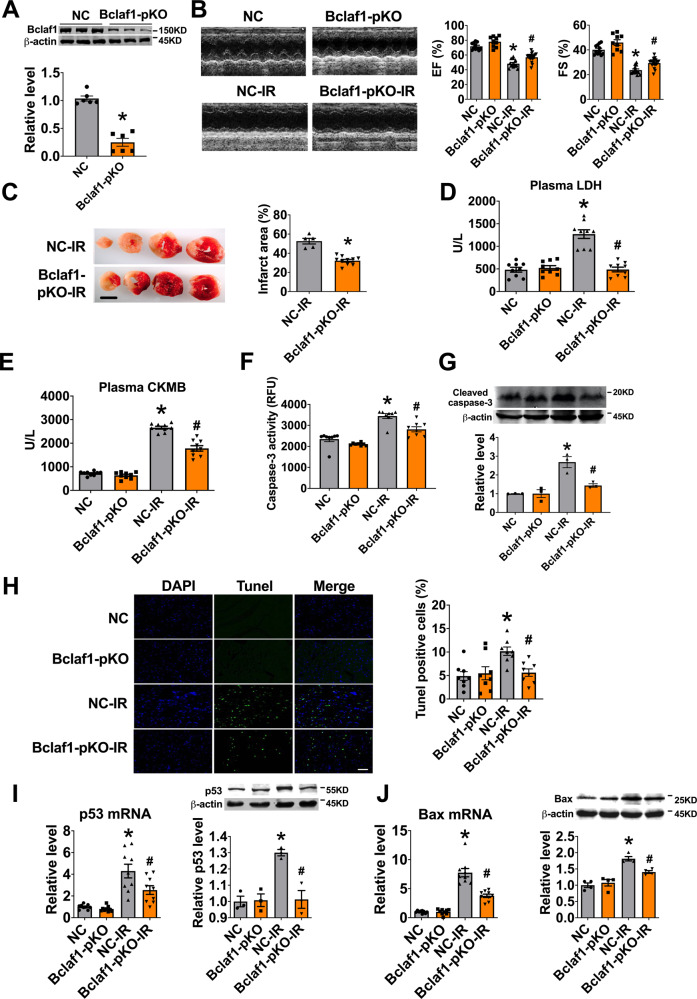
Fig. 5. Partial knockout (pKO) of Bclaf1 alleviates cardiac ischemia–reperfusion injury.
A Bclaf1 in the heart tissue after adeno-associated virus carrying cas9-Bclaf1 injection. N = 6. *P < 0.05 versus NC group. P values were determined by unpaired t test. B Representative images of echocardiographs and statistics of ejection fraction (EF) and fractional shortening (FS). NC N = 11, Bclaf1-pKO N = 9, NC-I/R N = 13, Bclaf1-pKO-I/R N = 18. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. C Infarct size by TTC staining (scale bar: 2 mm). NC-I/R N = 5, Bclaf1-pKO-I/R N = 10. *P < 0.05 versus NC-I/R, P values were determined by unpaired t test. D, E Serum concentrations of LDH and CKMB, respectively, measured by Elisa assay. N = 9 mice per group. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. F Caspase-3 activity. N = 8 mice per group. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. G Cleaved caspase-3 protein level. N = 3 mice per group. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. H Apoptosis of cardiomyocyte in the border zone (BZ) by TUNEL staining (scale bar: 20 μm). N = 8 mice per group. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. I The mRNA and protein levels of p53, respectively. N = 9 for mRNA and N = 3 for protein. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. J The mRNA and protein levels of Bax, respectively. N = 9 for mRNA and N = 4 for protein. *P < 0.05 versus NC, #P < 0.05 NC-IR. P values were determined by one-way ANOVA test followed by Bonferroni post hoc analysis. Data are expressed as mean ± SEM.