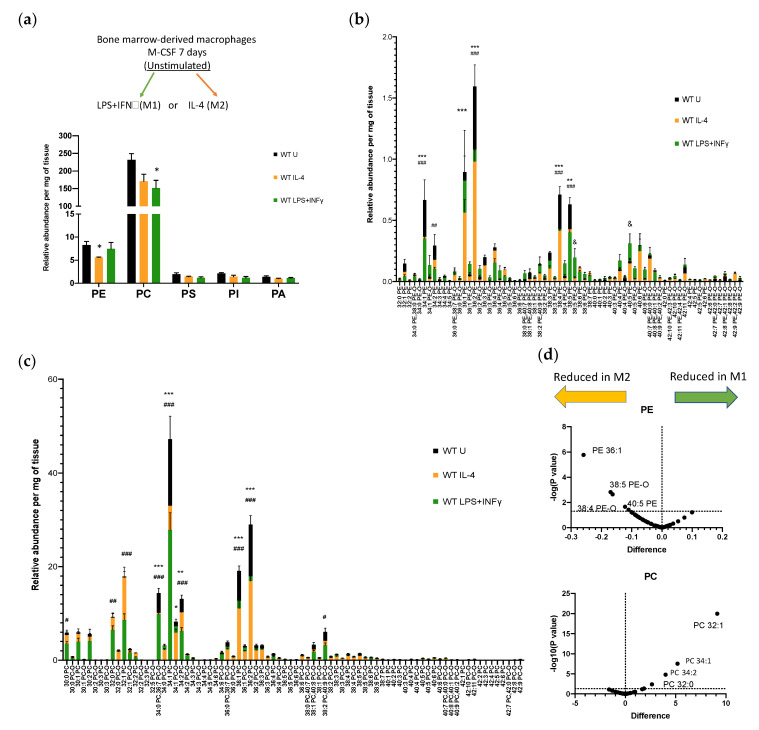
Figure 1.
Phospholipid metabolism in bone marrow-derived macrophages (BM-Macs). Wild Type (WT) BM-Macs were Unstimulated (U) or stimulated with Interleukin (IL)-4 or LPS+ Interferon (IFN)γ for 24 h, supernatants were removed and collected for analysis by liquid chromatography coupled to mass spectrometry (LC-MS). (a) * p < 0.05 of (U) vs. IL-4 or LPS+IFNγ BM-Macs. (b,d) Phosphatidylethanolamine (PE) molecules or (c,d) phosphatidylcholine (PC) molecules are identified by the binding of fatty acids which are described by the numbers of carbons and double bonds. PE-O and PC-O indicate ether linked fatty acids in the sn-1 position. The data are shown volcano plot of mean (d) or graph (a–c) of mean and standard error of three independent determinations. *** p < 0.0001, ** p < 0.005 (IL-4)BM-Macs reduction vs. (U)BM-Macs; ### p < 0.0001, ## p < 0.005, # p < 0.05 (LPS+IFNγ)BM-Macs reduction vs. (U)BM-Macs; & p < 0.05 increase in (LPS+IFNγ)BM-Macs vs. (U)BM-Macs. p-values were obtained by two-way ANOVA with Dunnett’s correction for multiple comparisons.