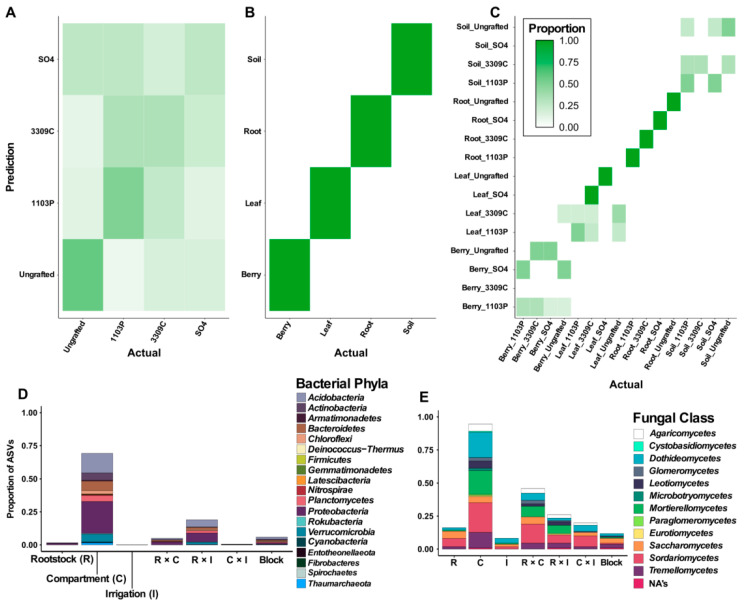
Figure 5.
Machine learning was able to distinguish between (A,B) the compartment from which the sample was collected but not rootstock genotype to which the vine was grafted and (C) when compartment and rootstock were jointly predicted only compartment was able to be predicted accurately. DESeq2 analysis found that multiple sources of variation were responsible for the patterns of differential abundance for amplicon sequence variants (ASVs) of (D) bacteria phyla and (E) Fungi classes.