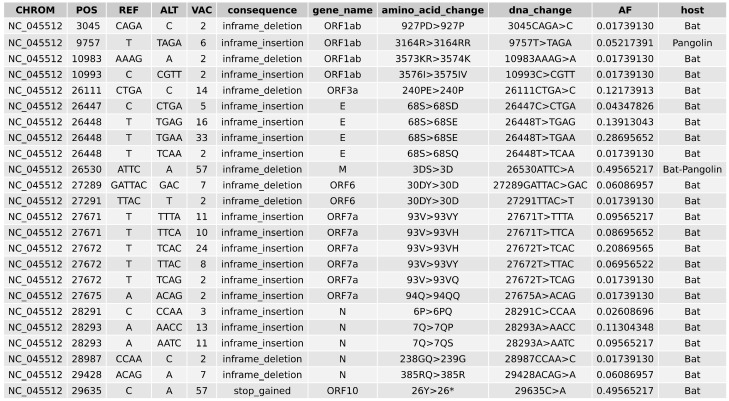
Figure 4.
High impact variants identified across bat and pangolin genomes using the variant calling pipeline based on SARS-Cov-2 Ensembl reference genome. The variants with allele frequency > 0.1 and predicted to have HIGH impact using VEPTools are listed. CHROM: Reference contig name; POS: Position; REF: Reference allele in Ensembl Human SARS-Cov2; ALT: Alternative allele(s) found in non-human genomes; VAC: Alternative variant allele counts; AF: Allele frequency.