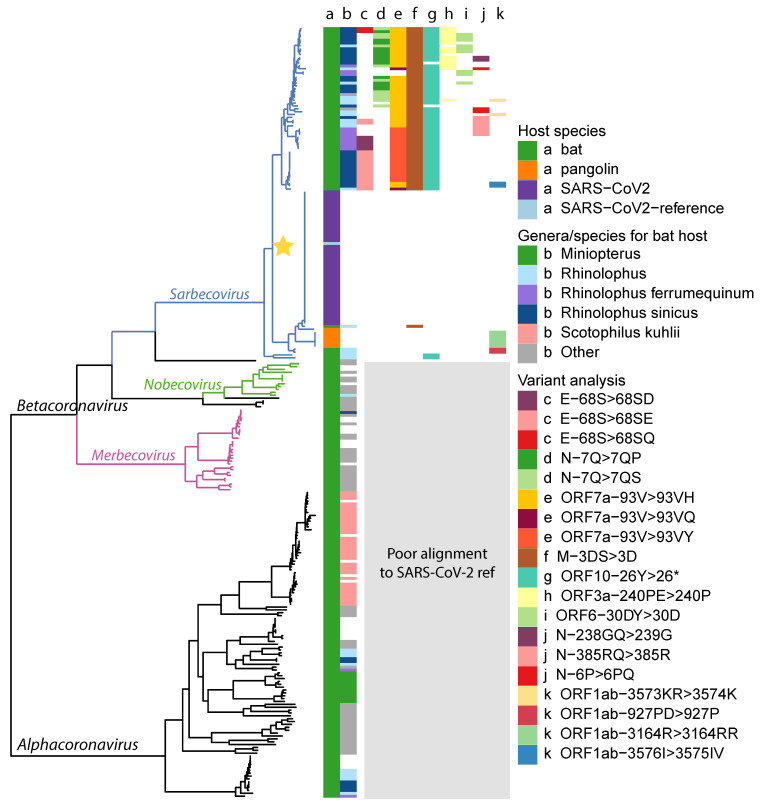
Figure A1.
Ladderised phylogenetic tree of bat-CoV, pangolin-CoV and SARS-CoV-2 (Wuhan dataset and reference) genomes. The hosts for each genome are indicated in (a) and host genera or species in (b) for bat-CoV. The majority of the Sarbecovirus affect the bat genus Rhinolophus (column b, light blue, dark blue and purple), whereas a much smaller proportion of the Alphacoronavirus are found in bats of this genus. Some clades overlap with specific bat species, including Rhinolophus ferrumequinum, Rhinolophus sinicus and Scotophilus kuhlii. Several high impact variants (inframe insertion, inframe deletion or stop gain) identified from variant analysis overlap with the clades in the phylogenetic tree. The annotation indicates (c–e) amino acid positions with multiple variants, (f–i) amino acid positions with a single change and found in >10 genomes, (k, l) other variants. The genes and amino acid changes involved in each of the annotated in-frame insertion, in-frame deletion or stop gain (*) are indicated in the figure legend. Star highlights the clade in Figure 1.