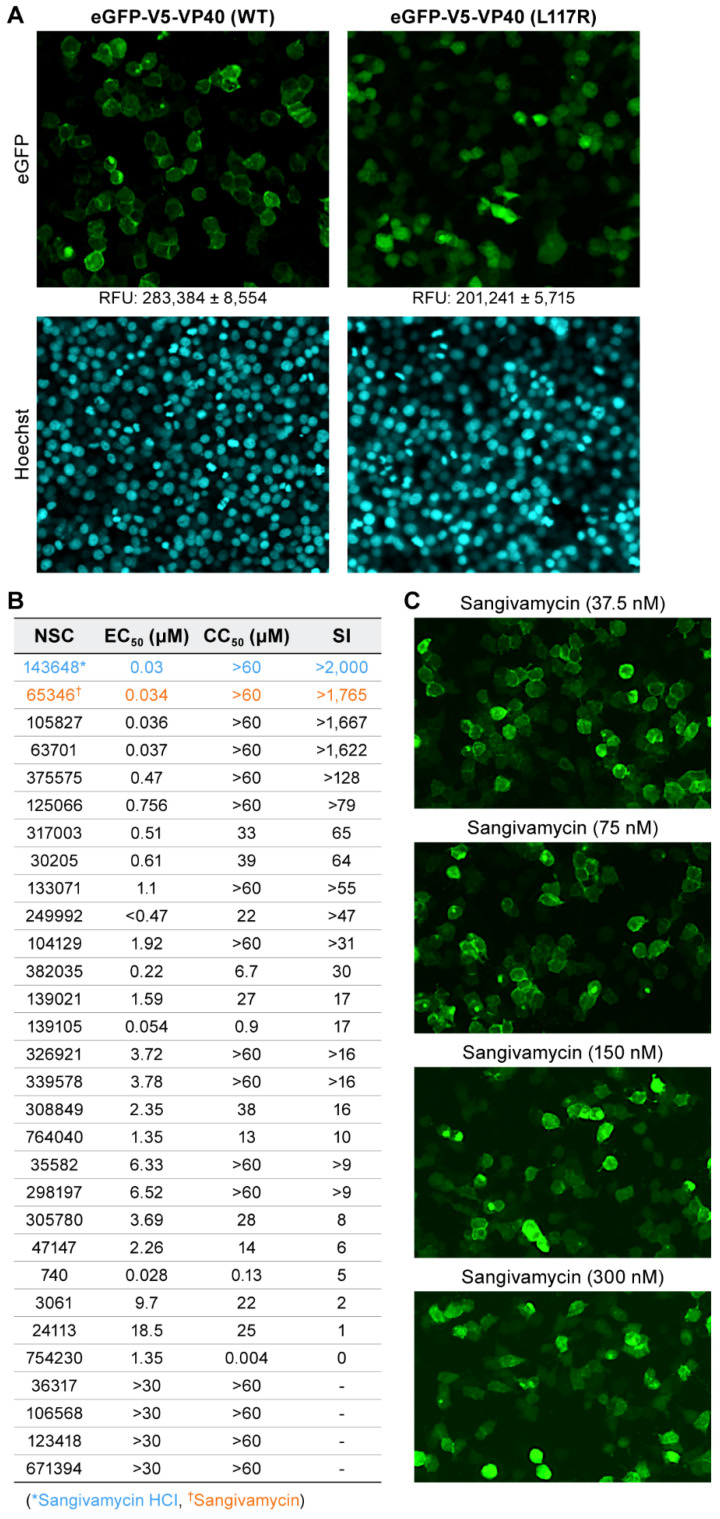
Figure 2.
EBOV VP40 membrane localization is disrupted by sangivamycin. (A) Disruption of VP40 membrane association via the introduction of the L117R mutation (VP40 L117R positive control). Relative fluorescence units (RFUs) are shown below the eGFP images at 10× magnification. Lower RFU values for hits were then phenotypically confirmed via VP40 membrane association and fluorescent cellular rings for VP40 WT (top left panel). VP40 L117R positive control prevents initial VP40 dimerization and virion-like particle (VLP) formation leading to diffuse cellular fluorescence with lower RFU (top right panel). Hoechst 33342 DNA staining for chromatin in nuclei was used as an initial indicator of cell health (bottom panels). (B) The table lists the top 30 hits by their NCI assigned NSC numbers. Sangivamycin as either its HCl salt (NSC 143648) or free base (NSC 65346) impairs EBOV VP40 localization to the inner plasma membrane. Listed are the half maximal effective concentration (EC50), drug concentration required to reduce cell viability by 50% (CC50), and selectivity index (SI) values of the 30 hits identified in the primary screening and evaluated in the secondary screening. EC50 values were quantified by plate reads compared to VP40 WT and VP40 L117R positive control values. CC50 values were measured with CellTiter-Glo as relative luciferase units compared to VP40 WT and VP40 L117R positive control and SI was calculated by CC50/EC50. Although initial quantification was performed by plate reader, all hits were confirmed by a phenotypic shift from fluorescent rings to homogenous expression by cellular imaging. (C) Representative fluorescent images of VP40 WT treated with 37.5, 75, 150, and 300 nM sangivamycin at 10× magnification.