Abstract
Epigenetic factors have been suggested as mediators of early-life nutrition to future health. Prior studies focused on breastfeeding effects on DNA methylation (DNAm), ignoring other feeding modes. In this analysis of the Isle of Wight birth cohort, feeding modes were categorized as exclusive breastfeeding (EBF), exclusive formula feeding (EFF), and mixed feeding based on whether the respective feeding mode lasted for at least 3 months. In addition, in the past, infant feeding modes were assessed using DNAm at one time point in childhood, not changes of DNAm. In this paper, methylation differences (delta DNAm) were calculated by subtracting residual methylation values at birth from age 10 years (adjusting for cell types and season of blood collection at both ages). These deltas were estimated for all methylation sites where cytosine was followed by guanine (cytosine guanine dinucleotide (CpG) sites). Then, we performed an epigenome-wide association study contrasting EBF, EFF, and mixed feeding with delta DNAm that represents changes in methylation from birth to 10 years. A total of 87 CpGs (EBF: 27 CpGs, EFF: 48 CpGs, mixed: 12 CpGs) were identified using separate linear regression models adjusting for confounders and multiple testing. The sum of all changes in methylation from birth to age 10 years was significantly lower in the EFF group. Correspondingly, the number of CpGs with a methylation decline was 4.7% higher reflecting 13,683 CpGs. Lower methylation related to exclusive formula feeding and its adverse potential for the child’s development needs future research to reduce adverse health effects.
Keywords: epigenome-wide association study, EWAS, epigenetics, DNA methylation, infant feeding, feeding mode, breastfeeding, formula feeding, mixed feeding, delta residuals
1. Introduction
Adequate nutrition throughout infancy and early childhood, especially during the first two years of life, is crucial for ensuring good health, optimal growth and development of newborns to their full potential [1,2]. Infant feeding practices include breast milk (BM) feeding and supplementary feeding (any food/liquid, including non-human milk or formula) [3]. Breast milk feeding can be distinguished into exclusive breastfeeding (BM along with vitamin/mineral supplements or medication), predominant breastfeeding (BM along with water, water-based drinks, fruit juice, vitamin/mineral supplements or medicine), and breastfeeding (BM along with any food/liquid, including non-human milk or formula) [3]. The World Health Organization (WHO) and several other organizations recommend exclusive breastfeeding for the first six months of life with the introduction of nutritionally adequate and safe complementary feeding starting from the age of six months with continued breastfeeding up to 2 years of age or beyond [4,5,6]. Despite infant formula trying to mimic the nutrition profile of human breast milk, BM stands out in terms of the colostrum/first fluid with unique immunologic components including anti-inflammatory, growth factors, and diverse gut microbiome [7,8,9]. Furthermore, the composition of human breast milk is dynamic as it changes over time within nursing sessions (foremilk and hindmilk), different stages of lactation (age of infant), and is surprisingly related to infant gender [10,11,12]. Compared to breastfed children, formula-fed children have higher risks of infectious morbidity, lower IQ scores, and are more likely to develop obesity, type 2 diabetes, and cardiovascular disease [13,14].
Epidemiological studies propose that epigenetics may constitute one of the potential mechanisms linking early life nutritional exposure with health and diseases in later life [15,16]. Epigenetics is the study of changes in gene activity that are mitotically heritable without changes in the DNA sequence. One of the most commonly studied epigenetic mechanisms related to early life nutrition is DNA methylation (DNAm)—the addition or removal of a methyl group most commonly at cytosine guanine dinucleotides (CpGs) [17,18]. DNAm has been proposed as a mechanism behind the long-term health effects of breastfeeding [19]. A recent systematic review suggests that findings from previous association studies on breastfeeding and DNAm are inconclusive and recommends more epigenome-wide association studies (EWASs) with proper measures of relevant covariates to be performed [20]. Our literature search identified ten association studies on breastfeeding and DNAm in humans (two candidate gene studies [21,22], two EWASs with breastfeeding as a covariate [23,24], and six EWASs with breastfeeding as an exposure variable [25,26,27,28,29,30]) (Table S1). It is important to note that these studies were highly diverse in terms of the age of assessment, tissue examined, array platform, DNAm targets and respective methods, and the categorization of breastfeeding. In addition, there is only one study that compared soy- and cow-based infant formula [31]. However, recent investigations on infant feeding and gene expression showed that infants’ current weight, height, and head circumference and expression of obesity-predisposing genes (FTO and CPT1A) are significantly higher in infants that were formula or mixed fed in comparison to exclusive breastfed [32]. Moreover, a mixed feeding mode (direct feeding at the breast, pumping and feeding, and formula feeding) was associated with a higher risk of food allergy symptoms than a single infant feeding source such as direct feeding at the breast and formula feeding [33].
Besides the limitations of prior association studies discussed in the recent systematic review [20], we identified two additional limitations in their study designs. First, most of the studies when assessing effects of breastfeeding duration on the epigenome were conducted irrespective of formula or solid feeding [25,26,27,28], which can be confounded by effects of breastfeeding. For example, infants could be mostly formula fed but still classified as breastfeeding in categorizations that have been widely investigated. To overcome this limitation, we used a more stringent categorization of infant feedings, such as exclusive breastfeeding, exclusive formula feeding and mixed feeding in our analysis. Second, past DNAm studies focused on measurement of DNAm at only one time point, e.g., 10 or 18 years [29], ignoring that the baseline of methylation can already be different at birth (before breastfeeding) and that DNAm at one point in childhood does not estimate changes that occur as a consequence of breastfeeding. Hence, only investigating the differences before and after breastfeeding, i.e., from birth to certain age, will provide appropriate assessments of epigenetic effects of infant feeding. Therefore, the objectives of this study are three-fold, (1) to examine whether DNAm changes from birth to ten years upon exposure to three infant feeding modes and if so, (2) to investigate the direction of association observed in each feeding group, and (3) to explore the effects of infant feeding on overall DNAm. Finally, we check whether the identified CpGs or genes were also reported in prior studies.
2. Materials and Methods
2.1. Study Setting and Participants
The data analyzed in this study are from the Isle of Wight Birth Cohort (IOWBC), the F1-generation (IoW F1). The IOWBC is a longitudinal cohort established to study the natural history of asthma and allergic conditions prospectively. The Isle of Wight is a small island (∼20 miles across) close to the British mainland with no industrial exposure and consists of primarily Caucasian participants (>98%). Potential study participants (n = 1536) were recruited between 1989 and 1990 by contacting parents (first generation, IoW F0) of all infants born on the IoW during this period, and approximately 95% of infants (n = 1456) were enrolled following informed consent and after exclusion of perinatal deaths, adoptions, moving, and refusals. Offspring were then followed at 1, 2, 4, 10, 18, and 26 years of age. Information about infant nutrition such as breastfeeding duration and introduction of formula and solids was gathered using questionnaires. The details on the study design, ethical approvals, enrollment, follow-up procedures, and variables assessed are described elsewhere [34,35]. Our analysis focused on the DNAm measurements at birth in DNA extracted from neonatal heel prick blood spots on Guthrie cards (n = 796), and at age 10 years (n = 330) in DNA from peripheral blood; 221 participants had DNAm data at both ages. After exclusion of missing information related to infant feeding, the analytical sample includes 201 participants.
2.2. Study Variables
2.2.1. Infant Feeding
Information on breastfeeding practices and total breastfeeding duration (relates to the number of weeks a mother breastfed her child regardless of the introduction of formula and/or solid food) and introduction of formula and solids were obtained through questionnaires answered by the mothers at the 1- and 2-year follow-up. For our analysis, we calculated exclusive breast feeding and exclusive formula feeding and mixed feeding duration (a combination of either of three feeding modes, breastfeeding/formula/solids) from the original feeding variables and used them as categorical variables. We used three months (13 weeks) as a cutoff point to categorize participants based on whether they were exclusively breastfed, exclusively formula fed, or a combination of any of the two feedings modes. Each feeding mode is determined as follows.
Exclusive Breastfeeding (EBF) group: child was only breastfed for at least 13 weeks of age before formula and/or solid foods were introduced.
Exclusive Formula Feeding (EFF) group: child was only formula fed (or cow’s milk) for at least 13 weeks of age (no breastfeeding) before solids were introduced.
Mixed feeding group: child was fed a mixture of breastmilk, formula, and solids before 13 weeks of age.
2.2.2. DNA Extraction and Profiling
DNA was extracted from the peripheral blood samples at age 10 (n = 330) using a standard salting-out procedure [36] and from dried heel blood samples (Guthrie cards) collected after the birth (n = 796) using a method based on the procedure described by Beyan et al. [37]. Approximately one microgram of DNA was bisulfite-treated for cytosine to thymine conversion using the EZ 96-DNA methylation kit (Zymo Research, Irvine, CA, USA) following the manufacturer’s standard protocol. Epigenome-wide DNAm was measured at birth and age 10 using Illumina BeadChip arrays (Illumina, Inc., San Diego, CA, USA). DNAm from Guthrie cards after birth were assessed using the Infinium Methylation EPIC BeadChip arrays measuring >850K CpGs. At age 10 years, both Infinium HumanMethylation450 BeadChip and EPIC BeadChip arrays were used and only CpG sites common between the two platforms were included in the analyses.
Methylation data were quantile normalized using the minfi R package and batch corrected using ComBat in R [38]. Probes not reaching a detection p-value of 10−16 in at least 95% of samples were excluded. To reduce the possible influence of probe single nucleotide polymorphisms (SNPs), CpG sites with probe SNPs within 10 base pairs and with minor allele frequency > 7% in the Caucasian population were further excluded. These steps resulted in 551,710 CpGs from 796 participants at birth, and 349,455 CpGs from 330 participants at age 10 years. Autosomal probes were then selected and converted to beta values, which represents the ratio of methylated (M) over the sum of methylated and unmethylated (U) probes (β = M/[c + M + U]), where c is used as a constant to prevent zero in the denominator. Finally, a total of 292,366 CpGs (present both at birth and age 10) were included in the statistical analyses.
2.2.3. Covariates
Variables known to potentially confound the association between duration of infant feeding and offspring DNAm levels were selected as covariates. These include categorical variables such as maternal smoking, birth order, socioeconomic status (SES), mode of delivery, and maternal asthma, and continuous variables such as birth weight and maternal age. Maternal age at delivery (in years) was calculated using child’s and maternal date of birth. Information about maternal smoking during pregnancy (question on active smoking) and maternal asthma (obtained from the questions of whether the mother suffered from asthma or wheezing attacks) were reported by the mothers at birth. Mode of delivery (recorded as Cesarean delivery, yes vs. no) and birthweight in kilograms were ascertained from the birth records. The birth order of the index child in the family (1st/2nd/3rd or more) was gathered by questionnaire. A composite family socioeconomic status (SES)-cluster variable that included a combination of parental occupation (reported at birth), number of children in a child’s bedroom (collected at age 4 years), and family income (at age 10) was used in our analyses. This composite variable has been described elsewhere [39].
In addition to covariates primarily related to breastfeeding, we also adjusted for variables that may confound effects of DNAm. These include infant’s sex [40], estimated cell-type composition of blood samples [41,42] and season of blood collection [43,44] at birth and age 10 years. Child’s sex was used to control the gender effects on the DNAm. We estimated proportion of cell types (CD8T, CD4T, natural killer cells, B Cells, monocytes, granulocytes) at each age (birth and 10 years) using the estimateCellCounts function from the minfi package [45] with an adult reference panel [42] following the Houseman approach [46]. Since Guthrie cards were collected between Day 3 and 5 after delivery, there was no need to additionally adjust the DNAm for nucleated red blood cells (nRBCs), as on Day 3 to Day 5 after birth nRBCs are no longer seen in the blood circulation of the newborn [47,48,49].
Season of blood collection at birth (obtained from the date of birth) and 10 years (obtained from the date of interview at 10 years of follow up) were regrouped as December–February (winter) March–May (spring), June–August (summer), and September–November (Fall) and used winter as reference [43].
2.3. Statistical Analysis
2.3.1. Comparison of the Analyzed Sample with the Complete Cohort
To evaluate whether the analyzed sample (subjects with DNAm data at birth and 10 years) differed from the study cohort, we compared the sample with the population using one-sample proportion tests (for categorical variables) and one-sample t-tests (for continuous variables).
2.3.2. Estimation of Differences of DNAm from Birth to Age 10 Years
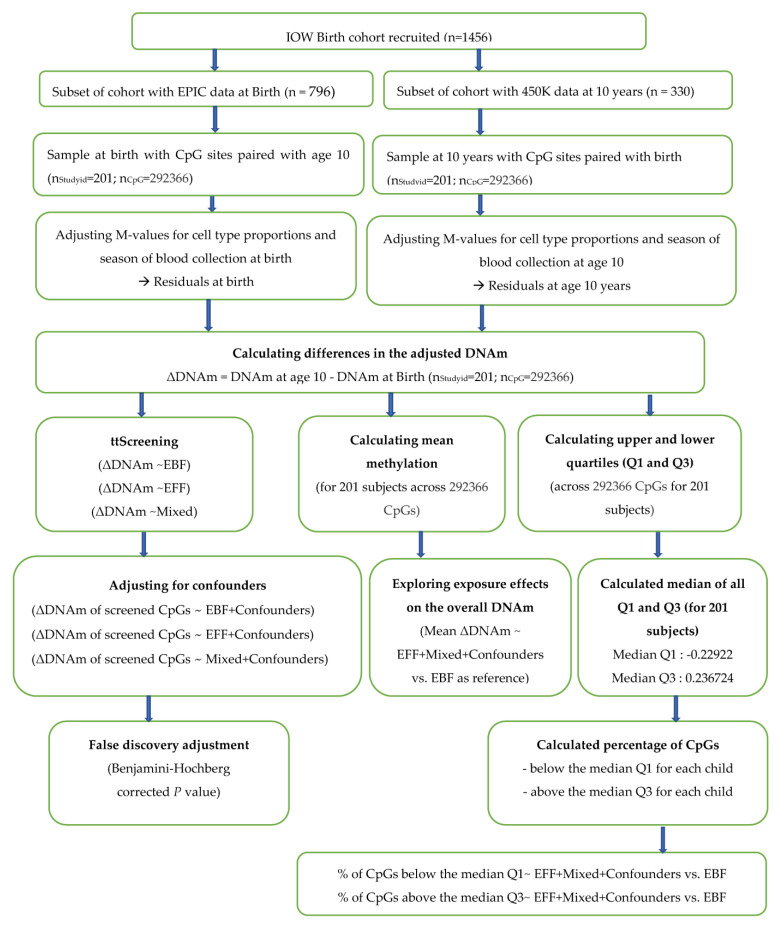
The analyses were restricted to paired data, i.e., CpGs and participants present both at birth and 10 years (292,366 CpGs and 201 participants). In the first step, to achieve normal distributions of DNAm measurements at birth and 10 years, we logit-transformed beta values to M values (using log2 (β value/(1-β value)). We then regressed M values on cell-type proportions and season of blood collection for each age (birth and 10 years) separately. This provides the residuals of DNAm (residDNAm) at birth and 10 years representing DNAm after correcting for cell types and season of blood collection. To determine differences in DNAm between birth and 10 years (delta or ‘Δ’ or delta-residuals (ΔDNAm)), the difference between the residuals at birth and 10 years was calculated (ΔDNAm = residDNAm at age 10–residDNAm at Birth). The delta-residuals (ΔDNAm) were treated as the outcome for further analysis.
2.3.3. Epigenome-Wide Discovery of Statistically Important CpGs Related to Infant Feeding
In the second step, we used delta-residuals, the changes in DNAm from birth to age 10 years, as an outcome to perform three separate EWASs for each infant feeding variable (EBF, EFF, and mixed) with two categories as the exposure by linear regression models. We screened all CpGs using the ttScreening R package, which was applied on delta-residuals to identify CpGs associated with each infant feeding variables separately using the following conceptual model:
| ΔDNAm(CpGi) = α + (β1 × feeding variable) |
where ΔDNAm(CpGi) denotes changes in DNAm at methylation site i and feeding variable represents one of the independent variables: EBF group, EFF group, and mixed group. This method employs training and testing data in robust linear regressions with surrogate variables in the regressions to adjust for unexplained variation in the data. The approach implemented in ttScreening was shown to perform better in controlling types I and II errors than the method controlling the false discovery rate (FDR) and the Bonferroni method [50]. CpGs that were not associated with infant feeding were excluded.
2.3.4. Linear Regression Adjusting for Confounder of the Discovered CpG Sites
To adjust for potential confounding, linear regression models were used for those CpGs that passed the screening with delta-residual at each CpG site as a dependent variable. All potential confounders (infant’s sex, cell compositions and season of blood collection at birth and age 10 years, maternal age at delivery, maternal smoking during pregnancy, mode of delivery, birthweight, birth order, and family socioeconomic status) were included in the model. Next, we compared the direction of significant regression coefficients across all three infant feeding modes using bar and volcano plots.
Additionally, we tested whether the overall global DNAm of 292,366 CpGs changed from birth to age 10 years. To this end, we first calculated the mean of the delta-residuals for each of the 201 subjects across all the 292,366 CpGs (paired data at birth and ten years). We then fitted a linear regression model using the means of all individual ΔDNAm as outcomes and the three infant feeding modes as exposures controlling for confounders (infant’s sex, cell compositions and season of blood collection at birth and age 10 years, maternal age at delivery, maternal smoking during pregnancy, mode of delivery, birthweight, birth order, and family socioeconomic status). Second, to investigate whether the overall DNAm was affected by infant feeding, we repeated the above equation with the average DNAm over all CpGs as outcome. Then, to test whether overall DNAm changes is due to minimal changes of all CpGs or higher changes in a limited proportion, we tested the proportion with positive (increase of DNAm) and negative changes (decrease in DNAm). To perform this, we first calculated the upper limit Q1 (lower quartile) and the lower limit Q3 (upper quartile) of all 292,366 CpGs for each of the 201 participants. Then, we determined the population medians of these Q1s and Q3s over all 201 children. For each child, we computed the percentage of CpGs below the median of the population-based Q1 (representing negative changes) and the corresponding percentage of CpGs above the median of the population-based Q3 (representing positive changes). Finally, we conducted two separate linear regression models using the percentages of CpGs (for 292,366 CpGs and 201 participants) that are below the population-based Q1 and the percentage of CpG that are above the Q3. Again, in these models, the three infant feeding modes were the exposures and we adjusted for confounders.
For all outcomes of the linear regression analyses, we checked the multi-variate normal distribution. The flow of the statistical analysis is shown in Figure 1. In all these analyses, multiple testing was adjusted by controlling a false discovery rate (FDR) of 0.05. Analyses were conducted using R package (version 3.6.1; Vienna, Austria) and SAS (version 9.4; SAS Institute, Cary, NC, USA).
Figure 1.
Flow chart of statistical analyses. nStudyid and nCpG represent the number of study participants and CpGs considered in the analysis, DNAm (DNA methylation), and Residual-DNA methylation after taking out the effects of cell-type proportions and season of blood collection at both time points. ttScreening—training and testing screening method, EBF—exclusively breastfed for at least 13 weeks, EFF—exclusively formula fed for at least 13 weeks, Mixed—a mixture of breastmilk with formula or solids before 13 weeks, ΔDNAm M-values—delta DNA methylation (calculated using M-values) that represents change in DNA methylation from birth to 10 years after adjusting for the cell-type proportions and season of blood collections at both time points.
2.4. Agreement of CpGs or Genes from Our Results with Prior Findings
In addition to statistical analysis, we performed a follow-up investigation of our findings to examine if we could find agreement between CpGs or genes from our analysis with those reported previously. Reports that used breastfeeding as a confounder, e.g., analyzed in tumor samples or studies that did not provide CpG sites information were excluded. The final list includes a total of 4308 CpGs, i.e., 12 CpGs from the study of Odintsova et al. [30], 13 CpGs from the study of Sherwood et al. [27,32], 7 CpG from Hartwig et al. [28], and 4276 CpGs from Naumova et al. [29]. We then used this list to look for the overlap of CpGs or their mapped genes with our findings.
2.5. Selective Screening of Associations with Continuous Health Outcomes
The scope of this paper does not allow for a systematic investigation of long-term health effects related to potential stable CpGs. However, within our study focusing on allergies and related factors, we tested the role of more stable CpGs for three outcomes with a continuous scale: one lung function test, serum IgE levels (a marker of allergy), and body mass index (BMI) at age 18. Measurements of these markers have been reported in prior papers [51,52,53].
Since health effects later in life require some stability of the DNAm changes, before linking Δ residuals of the DNAm to these three health outcomes, we tested how many of the discovered CpGs can be characterized as showing little dynamic in their methylation levels and thus could be classified as informative biomarkers in contrast to random effects. Using further information on DNAm at 18 years [54], we used mixed linear regression to check the temporal variability of the methylation of discovered CpGs between 10 and 18 years. To err on the safe side, we considered methylation changes between 10 and 18 years as dynamic, if there was a statistical indication of possible temporal changes with a p-value of 0.1 and no adjustment for multiple testing.
3. Results
3.1. Description of Study Participants
Among the 1456 enrolled children in the original cohort, 1360 were followed to age 10, of which 796 and 330 participants had DNA methylation available at birth and 10 years, respectively. For our analyses, we included 201 participants with methylation data for both ages and with non-missing information for infant feeding variables. The descriptive statistics comparing analyzed samples with the whole cohort are summarized in Table 1. The mean duration of total breastfeeding, the introduction of formula, and the introduction of solids were 14 weeks (±4.3), 8.9 weeks (±10.9), and 14.3 weeks (±4.6), respectively. The proportions of infants exclusively breastfed and formula fed for more than three months are 18.2% and 8.9%, respectively. Most infants (72.8%) received formula or solid foods before three months.
Table 1.
Comparing characteristics of the analyzed sample consisting of participants with DNA methylation data at both birth and age 10 with the initial Isle of Wight birth cohort.
| Variable | Total Cohort (N = 1456) | Participants with DNAm at Birth and Age 10 Year (N = 201) | |||
|---|---|---|---|---|---|
| N | n (%) | N | n (%) | p-Value | |
| Gender | |||||
| Male | 1456 | 748 (51.3) | 201 | 116 (57.7) | 0.07 |
| Maternal smoking | |||||
| Yes | 1442 | 362 (25.1) | 200 | 39 (19.5) | 0.07 |
| Missing | 14 | 1 | |||
| Birth order | |||||
| 1 | 1152 | 491 (42.6) | 188 | 80 (42.6) | 0.69 |
| 2 | 394 (34.2) | 60 (31.9) | |||
| 3 | 267 (23.2) | 48 (25.5) | |||
| Missing | 304 | 13 | |||
| Socioeconomic status | |||||
| High | 1293 | 104(8) | 201 | 21(10.5) | 0.30 |
| Medium | 995(76.9) | 115(77.1) | |||
| Low | 194(15) | 25(12.4) | |||
| Missing | 163 | 0 | |||
| Caesarean | |||||
| Yes | 1187 | 110 (9.3) | 167 | 19 (11.4) | 0.36 |
| Missing | 269 | 34 | |||
| Maternal asthma | |||||
| Yes | 1438 | 158 (10.9) | 200 | 25 (12.5) | 0.49 |
| Missing | 18 | 1 | |||
| Season of heel prick blood collection at birth | |||||
| Winter | 1455 | 417 (28.7) | 201 | 44 (21.9) | 0.03 |
| Spring | 364 (25) | 45 (22.4) | |||
| Summer | 352 (24) | 61 (30.4) | |||
| Fall | 322 (22) | 51 (25.4) | |||
| Missing | 1 | 0 | |||
| Season of blood collection at 10 years | |||||
| Winter | 1456 | 442 (30.4) | 201 | 66 (32.8) | 0.23 |
| Spring | 285 (19.6) | 29 (14.4) | |||
| Summer | 225 (15.4) | 37 (18.4) | |||
| Fall | 504 (34.6) | 69 (34.3) | |||
| Birth weight (Kilograms) | |||||
| Mean (SD) | 1432 | 3.4 (0.5) | 197 | 3.3 (0.6) | 0.19 |
| Missing | 24 | 4 | |||
| Maternal age at delivery (years) | |||||
| Mean (SD) | 1137 | 29.2 (12.1) | 184 | 29.6 (0.3) | <0.0001 |
| Missing | 319 | 17 | |||
| Duration of Breastfeeding (weeks) | |||||
| Mean (SD) | 1280 | 14.4 (14.7) | 201 | 15.3 (14.1) | 0.35 |
| Missing | 176 | 0 | |||
| Age at introduction of formula (weeks) | |||||
| Mean (SD) | 1304 | 8.9 (10.9) | 201 | 9.6 (10.5) | 0.34 |
| Missing | 152 | 0 | |||
| Age at introduction of Solids (weeks) | |||||
| Mean (SD) | 1252 | 14.3 (4.6) | 201 | 15.2 (5.2) | 0.01 |
| Missing | 204 | 0 | |||
| Exclusive breastfeeding (EBF) | |||||
| Yes | 1319 | 240 (18.2) | 201 | 46 (22.9) | 0.07 |
| Missing | 137 | 0 | |||
| Exclusive Formula feeding (EFF) | |||||
| Yes | 1343 | 119 (8.9) | 201 | 18 (8.9) | 0.98 |
| Missing | 113 | 0 | |||
| Mixed Feeding (Breastfeeding/Formula/Solids) | |||||
| Yes | 1318 | 956 (72.8) | 201 | 137 (68.2) | 0.14 |
| Missing | 138 | 0 | |||
The subsamples included in the study represented the complete cohort with respect to all exposure and confounding variables included in the analysis, except for season of heel prick blood collection at birth, maternal age at delivery, and age at the introduction of solids. Significant differences were observed in the proportion of children born during the spring (20% vs. 14%) and summer (15% vs. 18%) for season of blood collection at birth. The average maternal age (29.2 vs. 29.6) and age of solid food introduction in weeks (14.3 vs. 15.2) were slightly higher in the analytical sample than the whole cohort. All these variables were controlled for in our analyses as potential confounders.
3.2. EWAS of Infant Feeding Duration and Changes in DNA Methylation
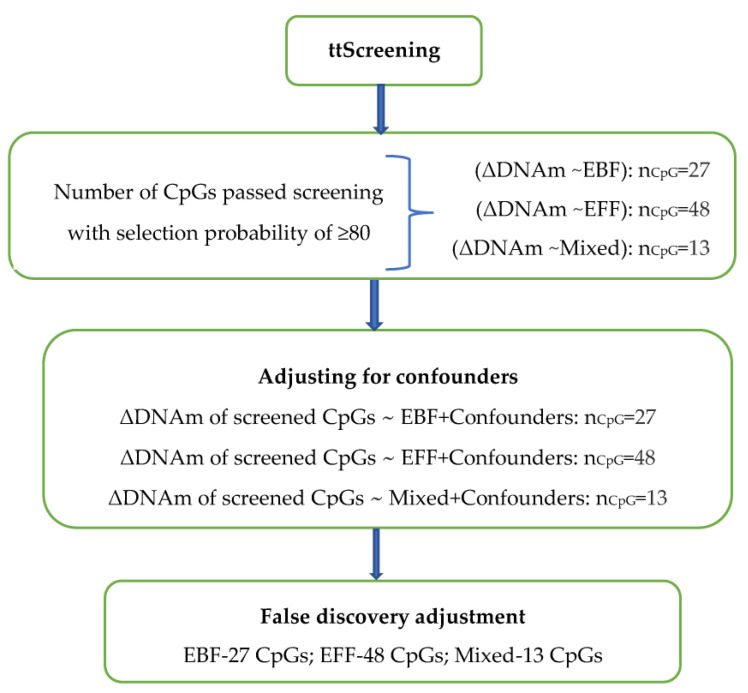
The training-testing method (ttScreening) analyzing 292,366 delta-residuals detected 87 informative CpGs across all the three feeding variables (27 CpGs for EBF, 48 CpGs for EFF, and 12 CpGs for mixed feeding) (Figure 2). The delta-residuals of these CpGs were further analyzed using linear regression models by controlling for confounders. After adjusting for multiple testing (FDR = 0.05), all CpGs remained significantly associated with three infant feeding variables (Table 2).
Figure 2.
Number of CpGs associated with each infant feeding variable at each stage of analysis. ttScreening—training-testing screening method, ΔDNAm—delta DNA methylation (calculated using M-values) that represents change in DNA methylation from birth to 10 years after adjusting for the cell-type proportions and season of blood collections at both time points, EBF—exclusively breastfed for at least 13 weeks, EFF—exclusively formula-fed for at least 13 weeks, Mixed—a mixture of breastmilk with formula or solids before 13 weeks. nCpG indicates the number of CpGs that are significant with a p-value of <0.05.
Table 2.
List of CpGs significantly associated with the different modes of infant feeding.
| Feeding Variable |
cgID | Coeff 1 | p-Value Raw | p-Value FDR2 | Gene Name | Chr 3 | CpG Location | Relation to UCSC 4 CpG Island |
|---|---|---|---|---|---|---|---|---|
| EBF | cg00367135 | 0.259 | 2.04 × 10−3 | 3.90 × 10−3 | BEST1 | 11 | Body | N_Shore |
| EBF | cg00825951 | 0.378 | 1.63 × 10−4 | 1.30 × 10−3 | RNF220 | 1 | 5′UTR | Island |
| EBF | cg00864821 | −0.292 | 4.75 × 10−4 | 1.80 × 10−3 | ITIH5; SFMBT2 | 10 | ||
| EBF | cg01153342 | 0.207 | 2.57 × 10−3 | 4.30 × 10−3 | C14orf43 | 14 | TSS1500 | S_Shore |
| EBF | cg01510332 | 0.204 | 8.10 × 10−3 | 9.50 × 10−3 | MGC2752 | 19 | Body | Island |
| EBF | cg01524359 | 0.245 | 3.66 × 10−3 | 5.80 × 10−3 | NASP | 1 | TSS1500 | N_Shore |
| EBF | cg01839850 | 0.319 | 3.14 × 10−4 | 1.70 × 10−3 | ANKRD54; MIR658 | 22 | TSS200 | Island |
| EBF | cg02418899 | 0.246 | 7.96 × 10−4 | 2.40 × 10−3 | ADCYAP1R1 | 7 | 5′UTR | Island |
| EBF | cg03607728 | 0.221 | 1.48 × 10−3 | 3.20 × 10−3 | RWDD2B | 21 | 1stE x on; 5′UTR | Island |
| EBF | cg05485520 | 0.237 | 1.31 × 10−3 | 3.20 × 10−3 | LZTFL1 | 3 | TSS200 | Island |
| EBF | cg05927802 | 0.275 | 6.12 × 10−4 | 2.10 × 10−3 | ENY2; NUDCD1 | 8 | Body; TSS1500 | Island |
| EBF | cg07303923 | −0.192 | 5.25 × 10−3 | 7.10 × 10−3 | TMEM217 | 6 | 5′UTR | N_Shelf |
| EBF | cg07399355 | 0.276 | 1.52 × 10−3 | 3.20 × 10−3 | MUC1 | 1 | TSS1500 | N_Shore |
| EBF | cg13585032 | 0.251 | 4.51 × 10−4 | 1.80 × 10−3 | ZNF786 | 7 | TSS1500 | S_Shore |
| EBF | cg13612447 | 0.179 | 1.90 × 10−2 | 1.97 × 10−2 | MIR1253 | 17 | Island | |
| EBF | cg13731777 | −0.253 | 5.13 × 10−3 | 7.10 × 10−3 | LGI1; SLC35G1 | 10 | ||
| EBF | cg14862791 | 0.208 | 7.42 × 10−3 | 9.10 × 10−3 | TTC23L | 5 | 5′UTR | Island |
| EBF | cg15415452 | 0.410 | 8.39 × 10−5 | 1.30 × 10−3 | SO X 1 | 13 | Island | |
| EBF | cg16924120 | 0.188 | 2.56 × 10−3 | 4.30 × 10−3 | SUDS3 | 12 | TSS1500 | Island |
| EBF | cg17393572 | 0.215 | 9.99 × 10−3 | 1.08 × 10−2 | HK3 | 5 | TSS200 | |
| EBF | cg17474435 | 0.191 | 9.36 × 10−3 | 1.05 × 10−2 | TMBIM6 | 12 | TSS200 | Island |
| EBF | cg17852997 | 0.284 | 1.51 × 10−3 | 3.20 × 10−3 | CRYL1 | 13 | TSS1500 | S_Shore |
| EBF | cg18462168 | 0.165 | 3.78 × 10−2 | 3.78 × 10−2 | CYFIP1 | 15 | TSS200 | Island |
| EBF | cg20357023 | 0.292 | 1.99 × 10−4 | 1.30 × 10−3 | TTC30B | 2 | TSS200 | Island |
| EBF | cg23266204 | 0.209 | 5.22 × 10−3 | 7.10 × 10−3 | EIF2B2 | 14 | 1stE x on | Island |
| EBF | cg25324653 | −0.300 | 1.11 × 10−4 | 1.30 × 10−3 | GHR; FB X O4 | 5 | ||
| EBF | cg25458520 | 0.164 | 6.60 × 10−3 | 8.50 × 10−3 | MAPK13 | 6 | Body | S_Shore |
| EFF | cg00993400 | −0.433 | 4.04 × 10−7 | <0.0001 | ZNF692 | 1 | TSS1500 | S_Shore |
| EFF | cg02351277 | −0.364 | 2.63 × 10−5 | <0.0001 | TBL1 X R1 | 3 | ||
| EFF | cg02396836 | −0.349 | 3.53 × 10−5 | 1.00 × 10−4 | MIR4255; ZC3H12A | 1 | N_Shelf | |
| EFF | cg03144848 | −0.364 | 4.05 × 10−5 | 1.00 × 10−4 | GPR20 | 8 | TSS200 | |
| EFF | cg04017131 | −0.449 | 1.94 × 10−3 | 2.00 × 10−3 | ANGPT2 MCPH1 | 8 | Body | |
| EFF | cg04587617 | −0.633 | 3.13 × 10−6 | <0.0001 | GRB7; IKZF3 | 17 | ||
| EFF | cg05613803 | −0.660 | 6.97 × 10−5 | 1.00 × 10−4 | KIAA0652 | 11 | Body; 1stE x on; 5′UTR; TSS200 |
Island |
| EFF | cg05629774 | 0.405 | 3.31 × 10−5 | 1.00 × 10−4 | EP400 | 12 | Body | |
| EFF | cg05697866 | −0.850 | 9.90 × 10−6 | <0.0001 | PLCH2; PANK4 | 1 | S_Shore | |
| EFF | cg07023208 | 0.340 | 7.99 × 10−5 | 1.00 × 10−4 | TAP2 | 6 | Body | |
| EFF | cg07168204 | 0.491 | 2.55 × 10−7 | <0.0001 | NDST1; RPS14 | 5 | N_Shore | |
| EFF | cg07470634 | −0.344 | 7.15 × 10−5 | 5.00 × 10−4 | P X N | 12 | 5′UTR; Body |
|
| EFF | cg08108289 | −0.752 | 6.97 × 10−9 | <0.0001 | ZBTB46; ABHD16B | 20 | Island | |
| EFF | cg08153335 | 0.291 | 4.44 × 10−4 | 5.00 × 10−4 | GPR113 | 2 | 5′UTR; 1stE x on; TSS200; Body | |
| EFF | cg08554115 | −0.434 | 2.76 × 10−5 | <0.0001 | LOC728392 | 17 | 1stE x on; 3′UTR | Island |
| EFF | cg08688798 | −0.742 | 1.09 × 10−5 | <0.0001 | DGCR6L | 22 | TSS1500 | Island |
| EFF | cg08704509 | −0.757 | 4.02 × 10−4 | 5.00 × 10−4 | GRAMD3 | 5 | Body; TSS200 |
N_Shore |
| EFF | cg09319125 | 0.377 | 2.05 × 10−4 | 3.00 × 10−4 | RBFO X 3; ENPP7 | 17 | ||
| EFF | cg09487373 | 0.399 | 2.14 × 10−4 | 3.00 × 10−4 | KCTD17; TMPRSS6 | 22 | N_Shelf | |
| EFF | cg10227331 | −0.358 | 4.10 × 10−4 | 5.00 × 10−4 | ERRFI1; PARK7 | 1 | ||
| EFF | cg10309425 | 0.305 | 4.63 × 10−4 | 5.00 × 10−4 | PRKCI; SKIL | 3 | ||
| EFF | cg10382845 | 0.371 | 6.12 × 10−5 | 1.00 × 10−4 | ABCC11 | 16 | Body | |
| EFF | cg11461670 | −0.579 | 1.14 × 10−6 | <0.0001 | PDLIM2 | 8 | 3′UTR | N_Shore |
| EFF | cg11597185 | 0.403 | 2.81 × 10−4 | 4.00 × 10−4 | DHCR7 | 11 | Body | N_Shelf |
| EFF | cg12106894 | 0.388 | 4.63 × 10−5 | 1.00 × 10−4 | KCNK6 | 19 | Body | S_Shelf |
| EFF | cg12491659 | −0.406 | 8.05 × 10−7 | <0.0001 | GAPT | 5 | TSS1500 | |
| EFF | cg14027297 | 0.374 | 1.61 × 10−5 | <0.0001 | TFIP11; TPST2 | 22 | S_Shelf | |
| EFF | cg14161438 | −0.509 | 1.60 × 10−5 | <0.0001 | ZADH2 | 18 | Body | Island |
| EFF | cg14398449 | −0.357 | 5.61 × 10−5 | 1.00 × 10−4 | POM121 | 7 | 5′UTR | N_Shore |
| EFF | cg14737571 | 0.512 | 2.90 × 10−5 | <0.0001 | ATP11A | 13 | Body | N_Shelf |
| EFF | cg15924285 | −0.470 | 9.09 × 10−5 | 2.00 × 10−4 |
SNORA8; SNORA1
SNORA32; SNORD6 |
11 | TSS200; TSS1500 |
|
| EFF | cg16746362 | 0.307 | 4.03 × 10−4 | 5.00 × 10−4 | C6orf25 | 6 | Body | S_Shore |
| EFF | cg18555555 | 0.334 | 4.36 × 10−4 | 5.00 × 10−4 | FABP7 | 6 | TSS1500 | |
| EFF | cg18990042 | −0.258 | 2.72 × 10−3 | 2.80 × 10−3 | COL2A1 | 12 | TSS1500 | Island |
| EFF | cg19334030 | −0.325 | 8.79 × 10−4 | 1.00 × 10−3 | DDAH2 | 6 | 5′UTR | Island |
| EFF | cg19957727 | −0.476 | 6.08 × 10−5 | 1.00 × 10−4 | TBCCD1; DNAJB11 | 3 | 1stE x on; 5′UTR; TSS200 | Island |
| EFF | cg20291674 | −0.464 | 4.97 × 10−5 | 1.00 × 10−4 | EZR | 6 | 5′UTR; TSS1500 |
Island |
| EFF | cg22881914 | −0.483 | 2.50 × 10−4 | 4.00 × 10−4 | NID2 | 14 | TSS1500 | Island |
| EFF | cg23528708 | −0.377 | 5.24 × 10−4 | 6.00 × 10−4 | CCDC90A | 6 | TSS200 | Island |
| EFF | cg23936463 | −0.470 | 6.33 × 10−6 | <0.0001 | FAM19A5 | 22 | TSS1500 | Island |
| EFF | cg24456654 | −0.414 | 3.10 × 10−4 | 4.00 × 10−4 | PHO X 2B; LIMCH1 | 4 | S_Shore | |
| EFF | cg24741888 | 0.278 | 1.62 × 10−4 | 3.00 × 10−4 | USP38; GAB1 | 4 | ||
| EFF | cg24975769 | −0.413 | 1.05 × 10−4 | 2.00 × 10−4 | F X C1; ARFIP2 | 11 | Body; TSS1500 |
S_Shore |
| EFF | cg25350011 | −0.526 | 1.54 × 10−3 | 1.60 × 10−3 | ANGPT2; MCPH1 | 8 | Body | |
| EFF | cg25687894 | −0.453 | 1.06 × 10−4 | 2.00 × 10−4 | ACLY | 17 | TSS1500 | S_Shore |
| EFF | cg25968437 | −0.243 | 3.06 × 10−3 | 3.00 × 10−3 | MLN; LEMD2 | 6 | ||
| EFF | cg26642844 | 0.283 | 5.99 × 10−4 | 7.00 × 10−4 | EIF3B | 7 | 3′UTR | N_Shore |
| EFF | cg27343836 | −0.279 | 5.23 × 10−5 | 1.00 × 10−4 | FO X F1 | 16 | Island | |
| Mi × ed | cg00809021 | 0.168 | 5.85 × 10−4 | 9.00 × 10−4 | CISH; MAPKAPK3 | 3 | ||
| Mi × ed | cg02165095 | 0.198 | 2.42 × 10−3 | 2.40 × 10−3 | ST8SIA6 | 10 | Body | |
| Mi × ed | cg02657836 | −0.284 | 4.33 × 10−4 | 7.00 × 10−4 | SGCD | 5 | S_Shore | |
| Mi × ed | cg03445694 | −0.244 | 1.34 × 10−3 | 1.50 × 10−3 | LH X 8 | 1 | Body | Island |
| Mi × ed | cg04086189 | 0.394 | 4.44 × 10−5 | 3.00 × 10−4 | SLF2; PA X 2 | 10 | ||
| Mi × ed | cg04797323 | 0.437 | 1.31 × 10−3 | 1.50 × 10−3 | SOCS2 | 12 | Body | Island |
| Mi × ed | cg06491101 | −0.311 | 3.65 × 10−4 | 7.00 × 10−4 | RICTOR | 5 | Body | |
| Mi × ed | cg06904618 | −0.172 | 1.63 × 10−4 | 4.00 × 10−4 | CT X N3; CT X N3 | 5 | Body | |
| Mi × ed | cg08600430 | 0.266 | 1.86 × 10−4 | 4.00 × 10−4 | SO X 1 | 13 | N_Shore | |
| Mi × ed | cg09412707 | 0.246 | 5.59 × 10−5 | 3.00 × 10−4 | SMIM20; SEL1L3 | 4 | ||
| Mi × ed | cg22976313 | 0.211 | 8.91 × 10−5 | 4.00 × 10−4 | TMEM179 | 14 | Body | N_Shore |
| Mi × ed | cg25324653 | 0.229 | 1.05 × 10−3 | 1.40 × 10−3 | GHR; FB X O4 | 5 |
EBF—Exclusively breastfed for at least 13 weeks. EFF—EFF-Exclusively formula fed for at least 13 weeks. Mixed—Given a mixture of breastmilk with formula/solids before 13 weeks. 1 Coeff: Regression coefficients represent changes in DNAm from birth to age 10 with each feeding variable. 2 FDR: False discovery rate. 3 Chr: Chromosome.4 UCSC: University of California Santa Cruz.
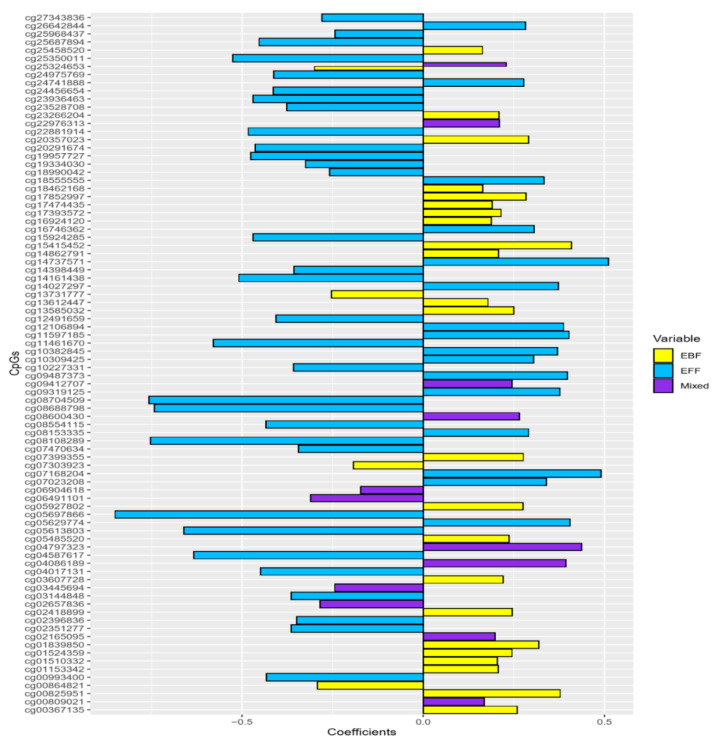
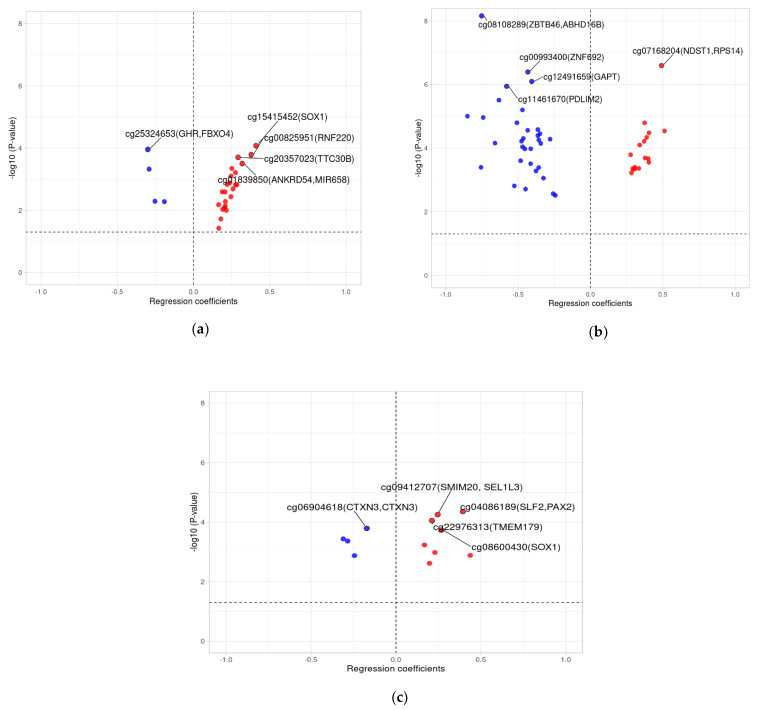
Inspecting the direction of regression coefficients in each infant feeding mode using bar and volcano plots shows that more CpGs related to EFF have negative regression coefficients (representing decreased methylation from birth to 10 years) than EBF and mixed feeding (Figure 3 and Figure 4).
Figure 3.
Bar plots of the regression coefficients by type of infant feeding mode. A negative coefficient represents methylation levels that are higher at birth and decreased at age 10 after exposure to different modes of feeding. A positive coefficient between ΔDNA methylation and feeding modes indicates that infant feeding is related to an increase in DNAm from birth to age 10.
Figure 4.
Volcano plots of −log10 (p-value) against regression coefficients of each infant feeding mode after adjusting for the confounders. The lower part of the volcano plot is not shown. The x-axis represents change of DNAm from birth to 10 years and the y-axis is −log10 transformed raw p-values (−log10p). The red-color dots denote positive regression coefficients and are towards the right side, and the blue-color dots represent negative coefficients are towards the left side of the plots. The horizontal dashed line indicates cutoffs for significance of raw p < 0.05, vertical dashed lines indicate regression coefficient cutoff of zero. (a) Volcano plot showing a total of 27 CpG sites methylated in the exclusive breastfeeding group with p < 0.05. (b) Volcano plot showing a total of 48 CpG sites methylated in the exclusive formula feeding group with p < 0.05. (c) Volcano plot showing a total of 12 CpG sites methylated in mixed feeding group with p < 0.05.
A linear regression model suggests that globally, over all 292,366 CpGs, the average DNAm declined more in exclusive formula-feed infants compared breastfed infants (Table 3). The mixed feeding mode showed no statistically significant association with the overall changes in DNAm between birth and 10 years of age.
Table 3.
Association of infant feeding modes and overall changes in DNA methylation based on 292,366 CpGs from birth to 10 years.
| Feeding Mode | Coeff 1 | Standard Error | p-Value |
|---|---|---|---|
| Exclusively formula fed for at least 13 weeks | −0.029 | 0.012 | 0.02 |
| Mixture of breastmilk with formula/solids before 13 weeks | −0.004 | 0.007 | 0.58 |
| Exclusively breastfed for at least 13 weeks | Reference |
1 Coeff: Regression coefficients represent change in overall mean DNAm of 292366 CpGs from birth to age 10 (ΔDNAm residuals controlling for cell types and season of blood collection at both time points) upon exposure to feeding modes (adjusting for confounders).
In addition, the percentage of CpGs with a more pronounced reduction in DNAm (ΔDNAm in the lower quartile of the distribution) was 4.7% (reflecting 13, 683 CpGs) higher in exclusive formula-feed compared to breast-fed infants controlling for confounders (Table 4). No associations were found for the mixed infant feeding mode and for the percentages of ΔDNAm increases over all CpGs. The average DNAm changes and the percentages of DNAm in the lower and upper quartile did not deviate from a multi-variate normal distribution.
Table 4.
Association of the percentages of CpGs that change between birth and age 10 years (delta-residuals) in the lower 25% (below Q1) and in the higher 25% distribution (above Q3) of the methylation of single CpGs related to infant feeding modes.
| Feeding Mode | Below Q1 1 (Lowest Quartile) |
Above Q3 2 (Highest Quartile) |
||
|---|---|---|---|---|
| Coeff 3 (in Percent) |
p-Value | Coeff 4 (in Percent) |
p-Value | |
| Exclusively formula fed for at least 13 weeks | 4.7 | 0.001 | −0.8 | 0.57 |
| Mixture of breastmilk with formula/solids before 13 weeks | 1.3 | 0.13 | 0.4 | 0.65 |
| Exclusively breastfed for at least 13 weeks | Reference | |||
1 Below Q1 represents percentages of CpGs that are lower than the Q1 level in the children. 2 Above Q3 represents percentages of CpGs that are higher than the Q3 level in the children. 3 Coeff is the regression coefficient that represents the difference of percentage of CpGs per participant that have delta-residuals below median Q1 cutoff comparing to the reference feeding model after adjusting for confounders (infant’s sex, cell compositions and season of blood collection at birth and age 10 years, maternal age at delivery, maternal smoking during pregnancy, mode of delivery, birthweight, birth order, and family socioeconomic status). 4 Coeff is the regression coefficient that represents the difference of percentage of CpGs per participant that has delta-residuals above median Q3 cutoff after adjusting for confounders (infant’s sex, cell compositions and season of blood collection at birth and age 10 years, maternal age at delivery, maternal smoking during pregnancy, mode of delivery, birthweight, birth order, and family socioeconomic status).
Interestingly the 87 CpGs discovered for exclusive breastfeeding (EBF), exclusive formula feeding (EFF), and mixed feeding were also statistically significantly differently distributed on locations on the genes (Table 5). The 27 CpGs related to EBF were more found in the transcription start site (48%), whereas the 48 CpGs related to EFF and the 12 CpGs linked to mixed feeding were found more in the body of the gene (35.4% and 50%, respectively).
Table 5.
Infant feeding mode and distribution of the CpG in different locations of the gene #.
| Exclusive Breast-Feeding (%) | Exclusive Formula Feeding (%) | Mixed Feeding (%) | Chi-Square Test, p-Value | |
|---|---|---|---|---|
| Transcription start sites (TSS) | 13 (48) | 17 (35) | 0 | 0.04 |
| Not TSS | 14 (52) | 31 (65) | 12 (100) | |
| Body of the gene incl. Exon 1 | 5 (18.5) | 17 (35.4) | 6 (50) | 0.009 |
| Not the body of the gene | 22 (81.5) | 31 (64.6) | 6 (50) |
# Whenever multiple locations on the gene were presented (e.g., Body, TSS1500), we counted both locations.
According to gene annotation, the 87 CpG sites having significantly different delta-residuals and associated with three feeding variables represent 117 genes. Of these 87 CpGs, we found only one common CpG (cg25324653; GHR, FBXO4) that was associated with both exclusive breastfeeding and mixed feeding (Table 2). We also discovered genes with more than one CpG linked to different feeding modes, i.e., SOX1 for cg15415452 (EBF; β = 0.409; P = 1.30 × 10−3) and cg08600430 (Mixed; β = 0.266; P = 4.00 × 10−4) and ANGPT2/MCPH1 for cg04017131 (EFF; β = −0.449; P = 2.00 × 10−3) and cg25350011 (EFF; β = −0.526; P = 1.60 × 10−3).
3.3. Agreement of CpGs and Genes Related to Infant Feeding from Prior Studies
We investigated the agreement of CpGs or genes that we identified with those reported in prior studies. Of the 4308 reported CpGs and their mapped genes (Table S2), we found no overlap in specific CpGs between our analysis with prior studies. However, we identified seven genes (RNF220, SFMBT2, LGI1, SUDS3, and CRYL1 associated with EBF; RBFOX3 and DDAH2 associated with EFF) from our study that were also reported previously (Table S3).
3.4. Associations with Continuous Health Outcomes
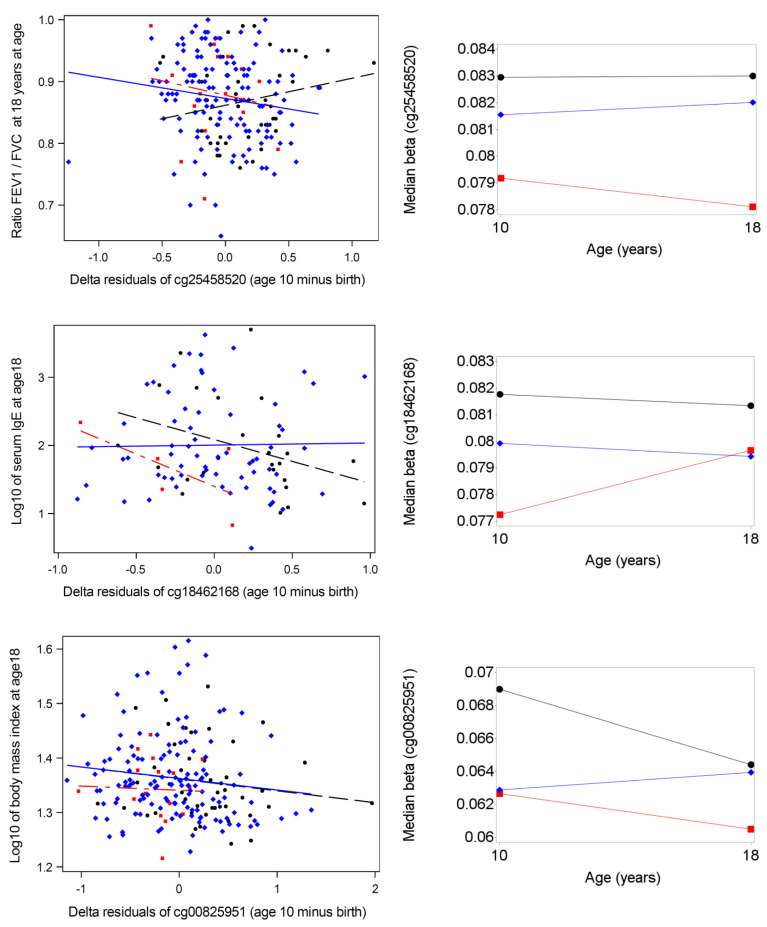
Testing the temporal variability between 10 and 18 years showed that 43 of the 86 CpG showed variability, but 43 CpG were more stable between 10 and 18 years, which nevertheless is a risky period with a number of changes related to puberty [55]. For the ratio of forced expiratory flow in 1 s and forced vital capacity (FEV1/FVC) Figure 5 (left site) shows that the increasing methylation of CpG site cg25458520 (MAPK13 gene) is related to an increase in FEV1/FVC if the child was exclusively breastfed (EBF) for 13 weeks, but to a reduction if the child was formula-fed (EFF) or received mixed feeding. However, the negative association was not as strong among formula-fed children, since the methylation levels were smaller. For immunoglobulin E (IgE, log10-scale), the figure shows a protective effect of both exclusive breastfeeding and formula feeding. Again, in EFF children, the protective effect did not reach the potential of EBF, since methylation levels of cg18462168 (CYFIP1 gene) in the EFF group were smaller. In the third example (Figure 5), higher methylation levels in all infant feeding groups were related to a smaller body mass index (BMI, log10 level). However, EFF children had smaller methylation and thus could not reach the potential of higher BMI reduction. More stable median levels in the three infant feeding groups are displayed on the right side of Figure 5. The Y-axis shows the proportion of methylation (ranging from 0 to 1). The CpGs sited were selected by excluding high variability in mixed linear models of repeated DNAm levels at ages 10 and 18 years.
Figure 5.
Scattergrams with regression lines for three different continuous health outcomes at age 18 years with delta residuals of three different CpGs and (left site) and medians of three CpGs at ages 10 and 18 years (N = 298) for three infant feeding groups: exclusive breastfeeding for 13 weeks—black, exclusive formula feeding for 13 weeks—red, mixed feeding mode—blue (right side). The ratio of forced expiratory flow in 1 s and forced vital capacity (FEV1/FVC) was based on lung function test at age 18, immunoglobulin E (IgE) was measured in serum in IU/mL, body mass index is defined as kg/m2.
4. Discussion
Infant nutrition is essential for growth, development, and overall future health. Although various infant feeding modes exist, most of the epigenetic studies focus only on the crude effect of breastfeeding on DNAm and at a single age in childhood. To our knowledge, this is the first EWAS to investigate the association between exclusive and mixed infant feeding modes and epigenome-wide change in DNAm from birth to 10 years. In the Isle of Wight Birth Cohort, we found that exclusive breastfeeding, exclusive formula feeding, and mixed feeding were significantly associated with DNAm at 27, 48, and 13 CpGs, respectively. The direction of DNAm changes (ΔDNAm) was surprising. Among the 87 identified CpG, we found a clear tendency for reduction of DNAm from birth to age 10 years for formula-fed infants. This motivated us to test overall changes. Considering the mean of changes of all 292,366 CpGs, formula-fed infants had a statistically significant lower average change, suggesting a reduction of their methylation levels at 10 years. Testing how many CpGs show a stronger reduction (lower quartile of the population-based distribution over all 292,366 CpGs) revealed that a statistically significant proportion of 4.7% of all CpGs (corresponding to 13,683 CpGs) are lowered from birth to age 10 years in formula-fed compared to breastfed children. This suggests that children who were given formula exclusively for at least three months had significantly lower methylation (both for the identified infant-feed mode associated CpGs and global DNAm changes) than children who were exclusively breastfed. In addition to the different direction of the methylation changes of different feeding modes, we also found that CpGs related to EFF and mixed feeding were more frequently found in the body of the gene, where CpGs related to EBF were often located in the transcription start site of the gene. It is possible that both the different direction of methylation changes and the location of the affected CpGs on the gene are associated. For instance, decreased in methylation and CpGs in the body of the gene may indicate a different genetic ‘programming’ [56]. However, these aspects cannot be covered in the scope of this paper but needs further analyses.
We considered the possible effects of most practiced infant feeding modes on DNAm changes by categorizing infants into three feeding groups. This is essential as it not only takes out the confounding effects of other infant feeding modes on breastfeeding while studying its association with DNAm, but also allows us to compare the effects of exclusive formula feeding and mixed feedings with recommended exclusive breastfeeding. Also, the use of these feeding modes is consistent with other association studies of infant feeding and adverse health outcomes such as obesity [57], gastroesophageal reflux [58], asthma [59], and food allergy [33]. About 18% of the infants in our cohort were exclusively breastfed for 13 weeks, 8.9% received formula exclusively for 13 weeks, and 73% received formula or solids (along with breastfeeding) before 13 weeks. Mixed feeding constituted the largest group. We used 13 weeks/3 months as a cutoff to categorize each of these three variables to provide sufficient sample size in each group for analysis.
Our statistical analysis used DNAm differences between birth and age 10 (M values of ΔDNAm or delta residuals) as an outcome by removing the effect of cell-type proportions and season of blood collection at both the time points. Therefore, a positive coefficient between infant feeding and delta methylation implies that DNAm was lower at birth and increased at 10 years following exposure to the infant feeding (EBF/EFF/mixed). On the other hand, a negative coefficient indicates that infant feeding is related to a decrease in DNAm from birth to 10 years. Note, we did not investigate differential DNAm detected in one measurement during childhood, but we did investigate changes of DNAm between birth and age 10 years for different infant feeding modes effective after birth. The use of these delta residual DNAm—compared to a single methylation assessment—is key to interpreting changes in methylation at age 10 with respect to baseline or birth, thus allowing us to examine the real exposure effects of different infant feeding modes on DNAm. For instance, as seen for the mother [60], the maternal wish to breastfeed may already result in a differential methylation at birth, which can be carried forward to childhood DNAm. However, this is not an effect of breastfeeding and would not be falsely detected when investigating changes in DNAm between birth and age 10 years.
Of the 86 discovered CpGs, we believe that 43 CpGs can be considered as potential biomarkers of infant feeding, since they show a significant effect when comparing DNAm between birth and 10 year and are more stable between 10 and 18 years.
A potential limitation is that we did not investigate differentially methylated regions (DMRs). There is a possibility that CpGs characterizing DMRs were not discovered by our approach. However, we also have to consider that DMR can be identified using the DNAm profiles in one assessment (at one age) focused either on exposures or diseases, but not for DNAm changes, which we addressed here. For instance, the literature does not suggest that changes in DNAm can be characterized by changes of methylation profiles covering specific genetic regions. In addition, there is a number of limitations involved in the detection and statistical modeling of DMRs [61]. Hence, focusing on CpG dinucleotide as biomarkers more easily facilitates future association studies between infant feeding modes and later health outcomes with CpG dinucleotides as potential mediators.
Another limitation is that, within the scope of this manuscript, we could not systematically cover the variety of all potential health outcomes that could be related to changes in DNAm. However, we showed potential associations with health outcomes later in childhood for a few continuous health indicators. The selection suggests that changes in DNAm between birth and age 10 may provide novel and interesting insights into later health effects and that changes in DNAm seems to constitute epigenetic biomarker that needs further evaluations.
Interestingly, infant feeding mode was not only related to specific DNAm but showed an overall wide-spread effect related to a significant decrease in methylation at 292,366 CpGs in exclusively formula-fed infants compared to exclusive breastfeeding. These unspecific consequences are in agreement with a non-specific variety of health outcomes related to infant feeding such as otitis media, lower respiratory tract and gastrointestinal infections, asthma, atopic dermatitis, childhood obesity, type 1 and type 2 diabetes, leukemia, sudden infant death syndrome, and overall infectious morbidity [7,8,9].
Although the current study is important for highlighting epigenetic modifications associated with different infant feeding modes, the following factors limit the conclusion of this investigation. Firstly, due to the small sample size in each feeding group and lack of formal replication for generating reproducible results, the findings of the study need to be considered with care. Second, while defining the breastfeeding group, we could not consider other common breastfeeding modes, such as direct feeding at the breast vs. pumping and feeding that have been shown to have different effects on other outcomes studied [35]. Furthermore, despite adjusting for a wide range of confounders in our analysis, there may still be residual confounding that cannot be eliminated due to unknown factors (psychosocial, environmental and nutritional factors related to lactation, etc.), which are not be controlled in the statistical analyses. Lastly, although the study reported reduced DNAm for exclusive formula feeding compared to exclusive breastfeeding, the underlying mechanisms remain unknown. Possible explanations may include either the direct effects of nutritional and bioactive molecules of breastmilk that are absent in formula, or indirectly by the gut microbiota that are impacted by formula, and by differences in the maternal nurturing behavior [62,63]. Also, another study on infant feeding revealed a high expression of FTO and CPT1A and a low expression of PPARA in formula-fed and mixed-fed groups compared to the exclusively breastfed fed group [32]. Hence, whether a decrease in methylation from birth to 10 years observed in case of exclusive formula feeding in the current study is also associated with differential gene expression and different disease risks has yet to be unraveled.
Future large-scale studies are warranted to evaluate the results of our study. It is crucial to investigate and determine etiologic mechanisms that mediate the observed associations between formula feeding and decreased methylation development up to ten years. Understanding the mechanisms related to this association will first help us to reduce the susceptibility of risks for adverse health outcomes in later life by optimizing infant nutrition. The mechanisms may include bioactive factors in breastmilk or the protective nurturing bond between mother and infant. Second, this information will guide us to develop novel interventions to improve children’s health.
Acknowledgments
We would like to acknowledge participation and cooperation of the children and parents of Isle of Wight and appreciate the hard work of Sharon Matthews and the Isle of Wight research team in collecting data and Nikki Graham for technical support. We are also thankful for the High-Performance Computing facility provided by the University of Memphis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6643/13/1/99/s1, Table S1. Overview of previous DNA methylation and breastfeeding studies; Table S2. List of all previously reported CpGs used to investigate overlapping CpGs; Table S3. List of all overlapped genes with prior studies, their corresponding CpGs, and associated feeding variable.
Author Contributions
Conceptualization, W.K., H.Z., Y.J., S.C. and N.M.; methodology, W.K., H.Z., Y.J., S.C. and N.M.; formal analysis, Y.M. and W.K.; investigation, S.H.A.; resources, W.K.; data curation, J.W.H., S.H.A. and S.E.; writing—original draft preparation, Y.M.; writing—review and editing, W.K., H.Z., Y.J. and N.M.; visualization, Y.M., W.K., Y.J., S.C. and N.M.; supervision, W.K., H.Z. and Y.J.; project administration, S.C. and N.M.; funding acquisition, W.K., H.Z., J.W.H., S.H.A. and S.E. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been supported by National Institute of Allergy and Infectious Diseases [R01 AI091905] and National Heart, Lung, and Blood Institute [R01 HL132321] to W.K. and by National Institute of Allergy and Infectious Diseases [R01AI121226, MPI: H Zhang and JW Holloway] and the National Asthma Campaign, UK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, USA, and National Asthma Campaign, UK.
Institutional Review Board Statement
The local research ethics committee (NRES Committee South Central—Hampshire B, UK) and University of Memphis Institutional Review Board in Memphis, U.S. (STUDY #: 2423) approved the IOW study.
Informed Consent Statement
At recruitment and at all follow-ups participants provided written consents.
Data Availability Statement
The data will be available on request from The David Hide Asthma and Allergy Research Centre, St Mary’s Hospital, Isle of Wight PO30 5TG, UK, attention Stephen Potter (ISLE OF WIGHT NHS TRUST) <stephenpotter1@nhs.net>.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bhutta Z.A., Ahmed T., Black R.E., Cousens S., Dewey K.G., Giugliani E.R.J., Haider B.A., Kirkwood B.R., Morris S.S., Sachdev H.P.S., et al. What works? Interventions for maternal and child undernutrition and survival. Lancet. 2008;371:417–440. doi: 10.1016/S0140-6736(07)61693-6. [DOI] [PubMed] [Google Scholar]
- 2.Schwarzenberg S.J., Georgieff M.K. Committee on Nutrition Advocacy for Improving Nutrition in the First 1000 Days to Support Childhood Development and Adult Health. Pediatrics. 2018;141:e20173716. doi: 10.1542/peds.2017-3716. [DOI] [PubMed] [Google Scholar]
- 3.WHO. IFPRI. UC Davis. USAID. FANTA. Macro International . Indicators for Assessing Infant and Young Child Feeding Practices. World Health Organization; Geneva, Switzerland: 2010. [Google Scholar]
- 4.WHO . The World Health Organization’s Infant Feeding Recommendation. World Health Organization; Geneva, Switzerland: 2002. [(accessed on 6 September 2020)]. Fifty-Fifth World Health Assembly. Available online: https://www.who.int/nutrition/topics/infantfeeding_recommendation/en/ [Google Scholar]
- 5.Beidelman A.I., Schanler R.J. Breastfeeding and the Use of Human Milk. Pediatrics. 2012;129:e827–e841. doi: 10.1542/peds.2011-3552. [DOI] [PubMed] [Google Scholar]
- 6.CDC . Strategies to Prevent Obesity and Other Chronic Diseases: The CDC Guide to Strategies to Support Breastfeeding Mothers and Babies. Department of Health and Human Services; Atlanta, GA, USA: 2013. [(accessed on 6 September 2020)]. Available online: https://www.cdc.gov/nutrition/InfantandToddlerNutrition/breastfeeding/recommendations-benefits.html. [Google Scholar]
- 7.Ballard O., Morrow A.L. Human Milk Composition. Pediatr. Clin. N. Am. 2013;60:49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin C.R., Ling P.-R., Blackburn G.L. Review of Infant Feeding: Key Features of Breast Milk and Infant Formula. Nutrients. 2016;8:279. doi: 10.3390/nu8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker W.A. Initial Intestinal Colonization in the Human Infant and Immune Homeostasis. Ann. Nutr. Metab. 2013;63(Suppl. S2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 10.Galante L., Milan A.M., Reynolds C.M., Cameron-Smith D., Vickers M.H., Pundir S. Sex-Specific Human Milk Composition: The Role of Infant Sex in Determining Early Life Nutrition. Nutrients. 2018;10:1194. doi: 10.3390/nu10091194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powe C.E., Knott C.D., Conklin-Brittain N. Infant sex predicts breast milk energy content. Am. J. Hum. Biol. 2010;22:50–54. doi: 10.1002/ajhb.20941. [DOI] [PubMed] [Google Scholar]
- 12.Van Sadelhoff J.H.J., Van De Heijning B.J.M., Stahl B., Amodio S., Rings E.H.H.M., Mearin M.L., Garssen J., Hartog A. Longitudinal Variation of Amino Acid Levels in Human Milk and Their Associations with Infant Gender. Nutrients. 2018;10:1233. doi: 10.3390/nu10091233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson S., Fall C.H. Infant Nutrition and Later Health: A Review of Current Evidence. Nutrients. 2012;4:859–874. doi: 10.3390/nu4080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuebe A. The risks of not breastfeeding for mothers and infants. Rev. Obstet. Gynecol. 2009;2:222–231. [PMC free article] [PubMed] [Google Scholar]
- 15.Canani R.B., Di Costanzo M., Leone L., Bedogni G., Brambilla P., Cianfarani S., Nobili V., Pietrobelli A., Agostoni C. Epigenetic mechanisms elicited by nutrition in early life. Nutr. Res. Rev. 2011;24:198–205. doi: 10.1017/S0954422411000102. [DOI] [PubMed] [Google Scholar]
- 16.Paparo L., Di Costanzo M., Di Scala C., Cosenza L., Leone L., Nocerino R., Canani R.B. The Influence of Early Life Nutrition on Epigenetic Regulatory Mechanisms of the Immune System. Nutrients. 2014;6:4706–4719. doi: 10.3390/nu6114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z.-X., Riggs A.D. DNA Methylation and Demethylation in Mammals. J. Biol. Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont C., Armant D.R., Brenner C.A. Epigenetics: Definition, Mechanisms and Clinical Perspective. Semin. Reprod. Med. 2009;27:351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verduci E., Banderali G., Barberi S., Radaelli G., Lops A., Betti F., Riva E., Giovannini M. Epigenetic Effects of Human Breast Milk. Nutrients. 2014;6:1711–1724. doi: 10.3390/nu6041711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartwig F.P., De Mola C.L., Davies N.M., Victora C.G., Relton C.L. Breastfeeding effects on DNA methylation in the offspring: A systematic literature review. PLoS ONE. 2017;12:e0173070. doi: 10.1371/journal.pone.0173070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obermann-Borst S.A., Eilers P.H., Tobi E.W., De Jong F.H., Slagboom P.E., Heijmans B.T., Steegers-Theunissen R.P. Duration of breastfeeding and gender are associated with methylation of the LEPTIN gene in very young children. Pediatr. Res. 2013;74:344–349. doi: 10.1038/pr.2013.95. [DOI] [PubMed] [Google Scholar]
- 22.Tao M.H., Marian C., Shields P.G., Potischman N., Nie J., Krishnan S.S., Berry D.L., Kallakury B.V., Ambrosone C., Edge S.B., et al. Exposures in early life: Associations with DNA promoter methylation in breast tumors. J. Dev. Orig. Health Dis. 2013;4:182–190. doi: 10.1017/S2040174412000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossnerova A., Tulupova E., Tabashidze N., Schmuczerova J., Dostal M., Rossner P., Gmuender H., Sram R.J. Factors affecting the 27K DNA methylation pattern in asthmatic and healthy children from locations with various environments. Mutat. Res. Mol. Mech. Mutagen. 2013;5:18–26. doi: 10.1016/j.mrfmmm.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Soto-Ramírez N., Arshad H., Holloway J., Zhang H., Schauberger E.M., Ewart S., Patil V.K., Karmaus W. The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clin. Epigenet. 2013;5:1. doi: 10.1186/1868-7083-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwig F.P., Smith G.D., Simpkin A.J., Victora C.G., Relton C., Caramaschi D. Association between Breastfeeding and DNA Methylation over the Life Course: Findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Nutrients. 2020;12:3309. doi: 10.3390/nu12113309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naumova O.Y., Odintsova V.V., Arincina I.A., Rychkov S.Y., Muhamedrahimov R.J., Shneider Y.V., Grosheva A.N., Zhukova O.V., Grigorenko E.L. A Study of the Association between Breastfeeding and DNA Methylation in Peripheral Blood Cells of Infants. Russ. J. Genet. 2019;55:749–755. doi: 10.1134/S1022795419060103. [DOI] [Google Scholar]
- 27.Odintsova V.V., Hagenbeek F.A., Suderman M., Caramaschi D., Van Beijsterveldt C.E.M., Kallsen N.A., Ehli E.A., Davies G.E., Sukhikh G.T., Fanos V., et al. DNA Methylation Signatures of Breastfeeding in Buccal Cells Collected in Mid-Childhood. Nutrients. 2019;11:2804. doi: 10.3390/nu11112804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pauwels S., Symons L., Vanautgaerden E.-L., Ghosh M., Duca R.C., Bekaert B., Freson K., Huybrechts I., Langie S.A., Koppen G., et al. The Influence of the Duration of Breastfeeding on the Infant’s Metabolic Epigenome. Nutrients. 2019;11:1408. doi: 10.3390/nu11061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherwood W.B., Bion V., Lockett G.A., Ziyab A.H., Soto-Ramírez N., Mukherjee N., Kurukulaaratchy R.J., Ewart S., Zhang H., Arshad S.H., et al. Duration of breastfeeding is associated with leptin (LEP) DNA methylation profiles and BMI in 10-year-old children. Clin. Epigenet. 2019;11:1–10. doi: 10.1186/s13148-019-0727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherwood W.B., Kothalawala D.M., Kadalayil L., Ewart S., Zhang H., Karmaus W., Arshad H., Holloway J., Rezwan F.I. Epigenome-Wide Association Study Reveals Duration of Breastfeeding Is Associated with Epigenetic Differences in Children. Int. J. Environ. Res. Public Health. 2020;17:3569. doi: 10.3390/ijerph17103569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harlid S., Adgent M., Jefferson W.N., Panduri V., Umbach D.M., Xu Z., Stallings V.A., Williams C.J., Rogan W.J., Taylor J.A. Soy Formula and Epigenetic Modifications: Analysis of Vaginal Epithelial Cells from Infant Girls in the IFED Study. Environ. Health Perspect. 2017;125:447–452. doi: 10.1289/EHP428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheshmeh S., Nachvak S.M., Rezvani N., Saber A. Effects of Breastfeeding and Formula Feeding on the Expression Level of FTO, CPT1A and PPAR-α Genes in Healthy Infants. Diabet. Metab. Syndr. Obes. Targets Ther. 2020;13:2227–2237. doi: 10.2147/DMSO.S252122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathias J.G., Zhang H., Soto-Ramirez N., Karmaus W. The association of infant feeding patterns with food allergy symptoms and food allergy in early childhood. Int. Breastfeed. J. 2019;14:1–12. doi: 10.1186/s13006-019-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arshad S.H., Holloway J.W., Karmaus W., Zhang H., Ewart S., Mansfield L., Matthews S., Hodgekiss C., Roberts G., Kurukulaaratchy R. Cohort Profile: The Isle of Wight Whole Population Birth Cohort (IOWBC) Int. J. Epidemiol. 2018;47:1043i–1044i. doi: 10.1093/ije/dyy023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arshad S.H., Karmaus W., Zhang H., Holloway J. Multigenerational cohorts in patients with asthma and allergy. J. Allergy Clin. Immunol. 2017;139:415–421. doi: 10.1016/j.jaci.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClelland M., Hanish J., Nelson M., Patel Y. KGB: A single buffer for all restriction endonucleases. Nucleic Acids Res. 1988;16:364. doi: 10.1093/nar/16.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyan H., Down T.A., Ramagopalan S.V., Uvebrant K., Nilsson A., Holland M.L., Gemma C., Giovannoni G., Boehm B.O., Ebers G.C., et al. Guthrie card methylomics identifies temporally stable epialleles that are present at birth in humans. Genome Res. 2012;22:2138–2145. doi: 10.1101/gr.134304.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 39.Ogbuanu I.U., Karmaus W., Arshad S.H., Kurukulaaratchy R.J., Ewart S. Effect of breastfeeding duration on lung function at age 10 years: A prospective birth cohort study. Thorax. 2009;64:62–66. doi: 10.1136/thx.2008.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singmann P., Shem-Tov D., Wahl S., Grallert H., Fiorito G., Shin S.-Y., Schramm K., Wolf P., Kunze S., Baran Y., et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenet. Chromatin. 2015;8:1–13. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adalsteinsson B.T., Gudnason H., Aspelund T., Harris T.B., Launer L.J., Eiriksdottir G., Smith A.V., Gudnason V. Heterogeneity in White Blood Cells Has Potential to Confound DNA Methylation Measurements. PLoS ONE. 2012;7:e46705. doi: 10.1371/journal.pone.0046705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.-E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lockett G.A., Soto-Ramírez N., Ray M.A., Everson T.M., Xu C.-J., Patil V.K., Terry W., Kaushal A., Rezwan F.I., Ewart S.L., et al. Association of season of birth with DNA methylation and allergic disease. Allergy. 2016;71:1314–1324. doi: 10.1111/all.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ricceri F., Trevisan M., Fiano V., Grasso C., Fasanelli F., Scoccianti C., De Marco L., Tos A.P.D., Vineis P., Sacerdote C. Seasonality Modifies Methylation Profiles in Healthy People. PLoS ONE. 2014;9:e106846. doi: 10.1371/journal.pone.0106846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermansen M.C. Nucleated red blood cells in the fetus and newborn. Arch. Dis. Child. Fetal Neonatal Ed. 2001;84:211F–215F. doi: 10.1136/fn.84.3.F211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kondo T., Sekizawa A., Saito H., Jimbo M., Sugito Y., Okai T. Fate of fetal nucleated erythrocytes circulating in maternal blood: Apoptosis is induced by maternal oxygen concentration. Clin. Chem. 2002;48:1618–1620. doi: 10.1093/clinchem/48.9.1618. [DOI] [PubMed] [Google Scholar]
- 49.Boskabadi H., Zakerihamidi M., Sadeghian M.H., Avan A., Ghayour-Mobarhan M., Ferns G.A. Nucleated red blood cells count as a prognostic biomarker in predicting the complications of asphyxia in neonates. J. Matern. Neonatal Med. 2017;30:2551–2556. doi: 10.1080/14767058.2016.1256988. [DOI] [PubMed] [Google Scholar]
- 50.Ray M.A., Tong X., Lockett G.A., Zhang H., Karmaus W.J.J. An Efficient Approach to Screening Epigenome-Wide Data. BioMed Res. Int. 2016;2016:1–16. doi: 10.1155/2016/2615348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziyab A.H., Karmaus W., Kurukulaaratchy R.J., Zhang H., Arshad S.H. Developmental trajectories of Body Mass Index from infancy to 18 years of age: Prenatal determinants and health consequences. J. Epidemiol. Community Health. 2014;68:934–941. doi: 10.1136/jech-2014-203808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karmaus W., Mukherjee N., Janjanam V.D., Chen S., Zhang H., Roberts G., Kurukulaaratchy R.J., Arshad H. Distinctive lung function trajectories from age 10 to 26 years in men and women and associated early life risk factors—A birth cohort study. Respir. Res. 2019;20:98. doi: 10.1186/s12931-019-1068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han L., Kaushal A., Zhang H., Perunthadambil Kadalayil L., Duan J., Holloway J., Karmaus W., Banerjee P., Tsai S.-F., Wen H.-J., et al. DNA methylation at birth is associated with childhood serum immunoglobulin e levels. Epigenet. Insights. 2020 doi: 10.1177/25168657211008108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S., Refaey H., Mukherjee N., Solatikia F., Jiang Y., Arshad S.H., Ewart S., Holloway J.W., Zhang H., Karmaus W. Age at onset of different pubertal signs in boys and girls and differential DNA methylation at age 10 and 18 years: An epigenome-wide follow-up study. Hum. Reprod. Open. 2020;2020:hoaa006. doi: 10.1093/hropen/hoaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han L., Zhang H., Kaushal A., Rezwan F.I., Kadalayil L., Karmaus W., Henderson A.J.W., Relton C., Ring S.M., Arshad H., et al. Changes in DNA methylation from pre- to post-adolescence are associated with pubertal exposures. Clin. Epigenetics. 2019;11:1–14. doi: 10.1186/s13148-019-0780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Vlodrop I.J.H., Niessen H.E.C., Derks S., Baldewijns M.M.L.L., Van Criekinge W., Herman J.G., Van Engeland M. Analysis of Promoter CpG Island Hypermethylation in Cancer: Location, Location, Location! Clin. Cancer Res. 2011;17:4225–4231. doi: 10.1158/1078-0432.CCR-10-3394. [DOI] [PubMed] [Google Scholar]
- 57.Patel N., Dalrymple K.V., Briley A.L., Pasupathy D., Seed P.T., Flynn A.C., Poston L., UPBEAT Consortium Mode of infant feeding, eating behaviour and anthropometry in infants at 6-months of age born to obese women—A secondary analysis of the UPBEAT trial. BMC Pregnancy Childbirth. 2018;18:1–11. doi: 10.1186/s12884-018-1995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen P.-L., Soto-Ramírez N., Zhang H., Karmaus W. Association between Infant Feeding Modes and Gastroesophageal Reflux: A Repeated Measurement Analysis of the Infant Feeding Practices Study II. J. Hum. Lact. 2017;33:267–277. doi: 10.1177/0890334416664711. [DOI] [PubMed] [Google Scholar]
- 59.Klopp A., Vehling L., Becker A.B., Subbarao P., Mandhane P.J., Turvey S.E., Lefebvre D.L., Sears M.R., Azad M.B., Daley D., et al. Modes of Infant Feeding and the Risk of Childhood Asthma: A Prospective Birth Cohort Study. J. Pediatr. 2017;190:192–199. doi: 10.1016/j.jpeds.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 60.Gruzieva O., Merid S.K., Chen S., Mukherjee N., Hedman A.M., Almqvist C., Andolf E., Jiang Y., Kere J., Scheynius A., et al. DNA Methylation Trajectories During Pregnancy. Epigenetics Insights. 2019;12:2516865719867090. doi: 10.1177/2516865719867090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suderman M., Staley J.R., French R., Arathimos R., Simpkin A., Tilling K. Dmrff: Identifying differentially methylated regions efficiently with power and control. bioRxiv. 2018:508556. doi: 10.1101/508556. [DOI] [Google Scholar]
- 62.Gabbianelli R., Bordoni L., Morano S., Calleja-Agius J., Lalor J.G. Nutri-Epigenetics and Gut Microbiota: How Birth Care, Bonding and Breastfeeding Can Influence and Be Influenced? Int. J. Mol. Sci. 2020;21:5032. doi: 10.3390/ijms21145032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lester B., Conradt E., Lagasse L.L., Tronick E.Z., Padbury J.F., Marsit C.J. Epigenetic Programming by Maternal Behavior in the Human Infant. Pediatrics. 2018;142:e20171890. doi: 10.1542/peds.2017-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be available on request from The David Hide Asthma and Allergy Research Centre, St Mary’s Hospital, Isle of Wight PO30 5TG, UK, attention Stephen Potter (ISLE OF WIGHT NHS TRUST) <stephenpotter1@nhs.net>.