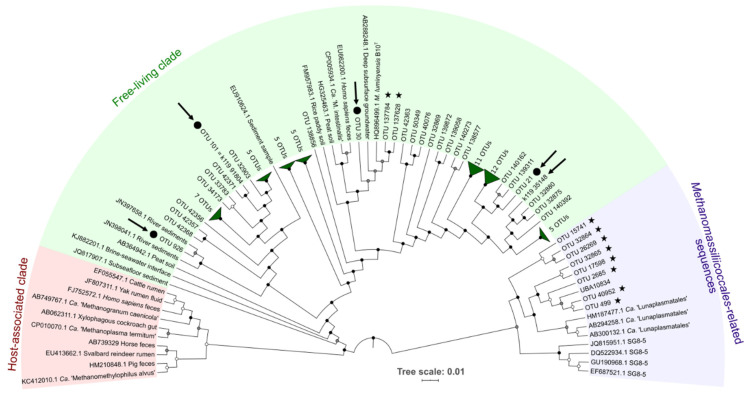
Figure 2.
Dendrogram showing the relationships between 16S rRNA gene sequences (253 bp) detected in environmental samples, in substrate-amended slurry (PENF slurry) and inferred to metagenome-assembled genomes (MAGs), among Methanomassiliicoccales [22]. The 85 OTUs of Methanomassiliicoccales detected in this study are shown on this dendrogram. OTUs detected only in environmental samples are indicated by a black star. OTUs without a symbol were detected in the culture-based experiment performed with the sediment PENF. OTUs indicated by a circle were detected both in bulk samples and in the culture-based experiment. The 16S rRNA gene sequences k119_35148 and k119_91804 were extracted from the metagenome sequences of the culture-based experiment PENF, after 8 weeks of incubation. Sequences discussed in the text are indicated by black arrows. The Methanomassiliicoccales-related sequences encompassed representative 16S rRNA gene sequences of Ca. ‘Lunaplasmatales’, UBA10834 and SG8-5 [80]. This reconstruction was performed on partial 16S rRNA gene sequences (253 bp), using the BIONJ method [81], with the modifications of Jukes and Cantor and using 1000 bootstrap replicates. Black-filled dots indicate nodes with bootstrap supports <50%, grey-filled dots 50–75%, and white-filled dots show support values between 75 and 100%. The top-right insert indicates the relative abundance of the main Methanomassiliicoccales-affiliated OTUs in the substrate-amended slurry PENF, after 8 weeks of incubation.