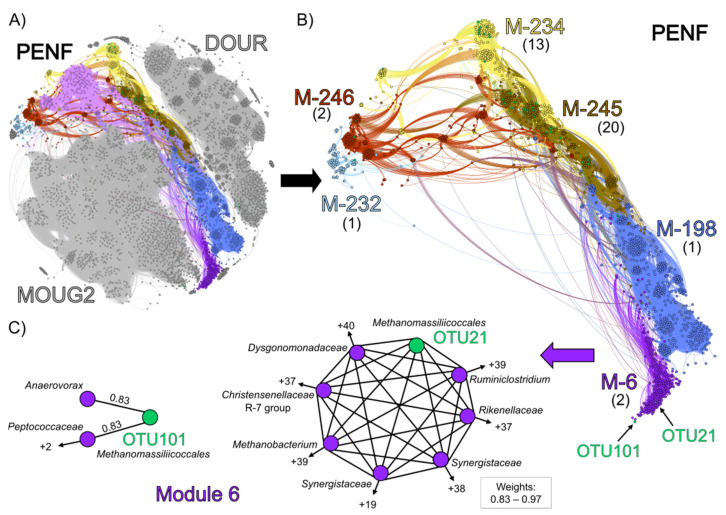
Figure 6.
Co-occurrence network reconstructed based on 16S rRNA gene-sequencing data from substrate-amended slurries (DOUR, PENF, and MOUG2, respectively), based on a Spearman rank’s correlation between OTUs and calculated with a percolation threshold of 0.79. (A) Clusters in slurries DOUR, PENF, and MOUG2, respectively. Modules in PENF network are colored to highlight them; (B) modules (M-6, module 6; M-198, module 198; M-232, module 232; M-234, module 234 and M-245, module 245, respectively) containing Methanomassiliicoccales in the cluster from the PENF culture-based incubation experiment. Modules consisting of a single Methanomassiliicoccales (modules 22 and 68, respectively) were excluded. The number of Methanomassiliicoccales in each module is shown in brackets; (C) non-random co-occurrences between Methanomassiliicoccales and other prokaryotes in module 6 showing co-occurrences of Methanomassiliicoccales (OTU21 and OTU101, respectively) with putative acetate- and H2-producing bacteria. Black arrows followed by a plus and a number indicate the number of taxa non-randomly co-occurring with other prokaryotes outside the groups represented. The weights of the co-occurrences are written above the link (OTU101) or in the insert below the cluster of the OTU21.