Correction to: Microbiome 7, 76 (2019)
https://doi.org/10.1186/s40168-019-0668-8
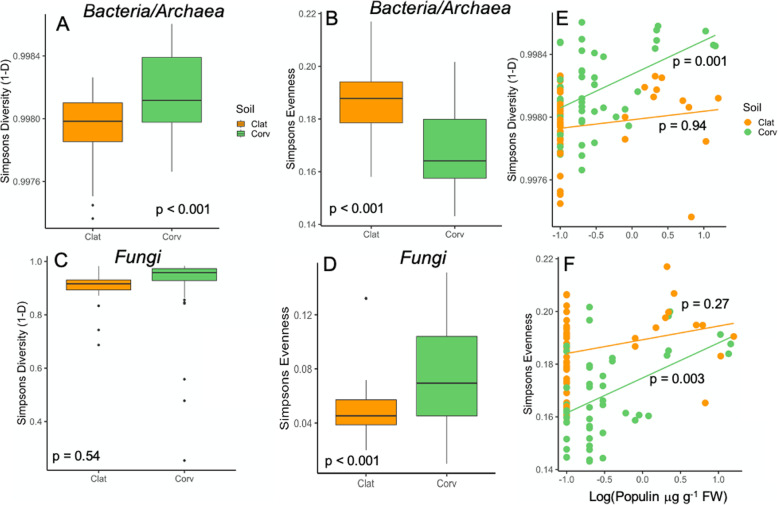
Following publication of the original article [1], the authors identified an error in Fig. 2. The correct figure is given below.
Fig. 2.
Bacterial/archaeal and fungal diversity (Simpson’s Diversity: 1-D; Panel a, c) and Simpsons’s Evenness (Panel b, d) in Clatskanie and Corvallis soil origins. Orange boxplots and points denote Clatskanie and green denotes Corvallis soils. Bacterial/archeal diversity and evenness was correlated with populin concentration in Corvallis soils (Panel e, f). Type-1 error rates given were generated by stepwise regression model analyses
Reference
- 1.Veach AM, Morris R, Yip DZ, et al. Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome. 2019;7:76. doi: 10.1186/s40168-019-0668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]